Plus Two Zoology Notes Chapter 4 Molecular Basis of Inheritance is part of Plus Two Zoology Notes. Here we have given Plus Two Zoology Notes Chapter 4 Molecular Basis of Inheritance.
| Board | SCERT, Kerala |
| Text Book | NCERT Based |
| Class | Plus Two |
| Subject | Zoology Notes |
| Chapter | Chapter 4 |
| Chapter Name | Molecular Basis of Inheritance |
| Category | Plus Two Kerala |
Kerala Plus Two Zoology Notes Chapter 4 Molecular Basis of Inheritance
The DNA
DNA is a long nucleotides polymer of deoxyribonucleotides.
1. Structure of Polynucleotide Chain:
A nucleotide has three components
- Nitrogenous base
- Pentose sugar (ribose in case of RNA, and deoxyribose for DNA), and
- Phosphate group.
There are two types of nitrogenous bases.
| Purines | Adenine and Guanine |
| Pyrimidines | Cytosine, Uracil and Thymine |
Cytosine is common for both DNA and RNA and Thymine is present in DNA.
Uracil is present in RNA at the place of Thymine.
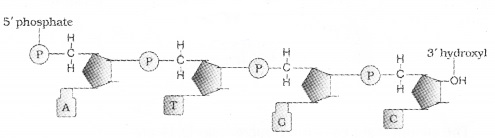
A nitrogenous base is linked to the pentose sugar through a N – glycosidic linkage to form a nucleoside, When a phosphate group is linked to 5′-OH of a nucleoside through phosphor ester linkage, to form nucleotide. Two nucleotides are linked through 3′-5′ phosphodiester linkage to form a dinucleotide.
Polynucleotide chain has at one end a free phosphate moiety at 5′-end of ribose sugar and other end of the polynucleotide chin the ribose has a free 3′-OH group The backbone in a polynucleotide chain is formed due to sugar and phosphates.
RNA has an additional-OH group present at 2′-position in the ribose and the uracil is found at the place of thymine.
Acidic nature of DNA was first identified by Friedrich Meischer in 1869 and it called as ‘Nuclein’.
| Data from the X-ray diffraction studies conducted by James Watson and Francis Crick, Maurice Wilkins and Rosalind Franklin showed that DNA has Double Helix structure. |
| Erwin Chargaff showed that ratios between Adenine and Thymine and Guanine and Cytosine are constant and equals one |
Each strand from a DNA acts as a template for synthesis of a new strand. The two double stranded DNA thus, produced would be identical to the parental DNA molecule.
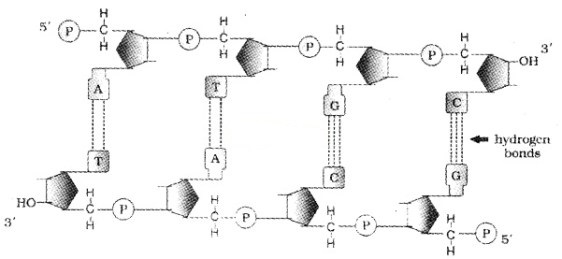
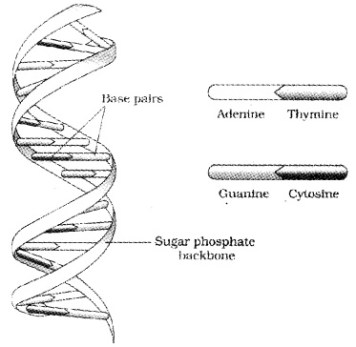
The salient features of the Double-helix DNA are
| (i) It is made of two polynucleotide chains, where the backbone is sugar-phosphate, (ii) The two chains have anti-parallel polarity, ie. 51 and 31 strands. (iii) The bases in two strands are paired through hydrogen bond. Adenine forms two hydrogen bonds with Thymine and Guanine is bonded with Cytosine with three H- bonds. (iv) The two chains are coiled in a right-handed fashion. The pitch of the helix is 3.4 nm. (v) The plane of one base pair stacks over the other in double helix. |
Francis Crick proposed the Central dogma in molecular biology, which states that the genetic information flows from DNA to RNA and RNA to protein. But in some viruses the flow of information is reverse direction, that is from RNA to DNA.
2. Packaging of DNA Helix:
In E. coli, they do not have a defined nucleus it is termed as ‘nucleoid’. The DNA in nucleoid is organised in large loops held by proteins.
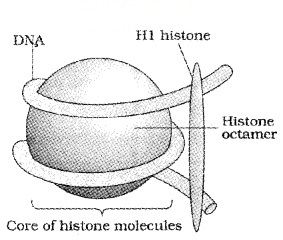
In eukaryotes, this organisation is much more complex. Here the positively charged, basic proteins called histones are associated with DNA.
Histones are rich in the basic amino acid residues lysines and arginines. Histones are organised to form a unit of eight molecules called as histone octamer.
The beads-on-string structure in chromatin is packaged to form chromatin fibers that are further coiled and condensed at metaphase stage of cell division to form chromosomes.
The negatively charged DNA is wrapped around the positively charged histone octamer to form a structure called Nucleosome. It contains 200 bp of DNA helix.
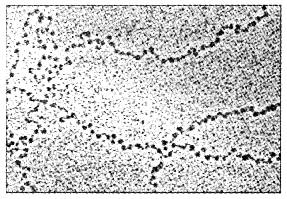
Nucleosomes constitute the repeating unit of a structure in nucleus called chromatin, it is thread-like stained (coloured) bodies seen in nucleus. The nucleosomes in chromatin are seen as ‘beads-on- string’ structure when viewed under electron microscope (EM).
The packaging of chromatin at higher level with proteins that are called as Non-histone Chromosomal (NHC) proteins.
| In a typical nucleus, some region of chromatin are loosely packed (euchromatin) and more densely packed (Heterochromatin). Euchromatin is transcriptionally active chromatin, whereas heterochromatin is inactive. |
The Search For Genetic Material
This is the work of identification of DNA that acts as a genetic material and responsible for inheritance.
Transforming Principle:
In 1928, Frederick Griffith, in a series of experiments with Streptococcus pneumoniae (bacterium responsible for pneumonia), showed the trasformation in the bacteria.
| For this, Streptococcus pneumoniae (pneumococcus) bacteria are grown, on a culture plate, some produce smooth shiny colonies (S) (mucous polysaccharide coat) while others produce rough colonies (R). |
Mice infected with the S strain (virulent) die from pneumonia infection but mice infected with the R strain do not develop pneumonia.
When Griffith was injected heat-killed S strain into mice, bacteria did not kill them. But he injected a mixture of heat-killed S and live R bacteria, the mice died and he recovered living S bacteria from the dead mice.
Biochemical Characterisation of Transforming Principle:
Oswald Avery, Colin MacLeod, and Maclyn McCarty worked on the ‘transforming principle’ of Griffith’s experiment and noticed that DNA alone from S bacteria caused R bacteria to become transformed.
They also discovered that protein-digesting enzymes (proteases) and RNA-digesting enzymes (RNases) did not affect transformation.
But the digestion with DNase inhibited transformation.
| They concluded that the transforming substance was not a protein or RNA but DNA is the hereditary material. |
1. Genetic Material is DNA:
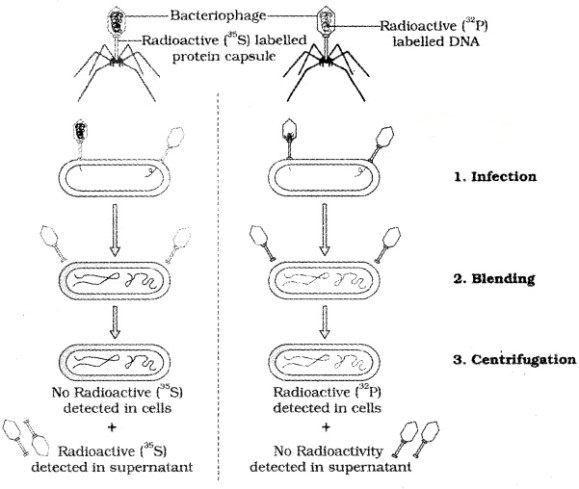
Alfred Hershey and Martha Chase (1952) grew bacteriophages on a medium that contained radioactive phosphorus and some others on medium that contained radioactive sulfur.
Viruses grown in the presence of radioactive phosphorus contained radioactive DNA but not radioactive protein because DNA contains phosphorus but protein does not. Similarly, viruses grown on radioactive sulfur contained radioactive protein but not radioactive DNA because DNA does not contain sulfur.
After the infection the viral coats were removed from the bacteria by agitating them in a blender. It is concluded that proteins did not enter the bacteria from the viruses. But the DNA is the genetic material that passed from virus to bacteria.
The Hershey-Chase experiment:
2. Properties of Genetic Material (DNA versus RNA):
A molecule that act as a genetic material must possess the following features
| (i) It should be able to generate its replica (Replication). (ii) It should chemically and structurally be stable. (iii) It should provide the scope for slow changes (mutation) that are required for evolution. (iv) It should be able to express itself in the form of ‘Mendelian Characters’. |
| The 2-OH group present at the nucleotide in RNA is a reactive group and makes RNA labile and easily degradable. Therefore, DNA is less reactive and structurally more stable when compared to RNA. |
Therefore, among the two nucleic acids, the DNA is a better genetic material.
The presence of thymine at the place of uracil also gives additional stability to DNA.
In fact, RNA being unstable, mutate at a faster rate. So the viruses having RNA genome having shorter life span mutate and evolve faster.
Rna World
RNA is genetic material as well as a catalyst. But it is reactive and hence unstable. Therefore, DNA has evolved from RNA with chemical modifications that make it more stable.
Replication
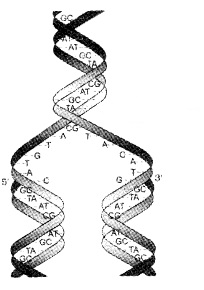
Watson and Crick (1953) proposed replication of DNA. They suggested that the two strands separate and act as a template for the synthesis of new complementary strands.
Waste-click model for semiconservative DNA replication:
1. The Experimental Proof:
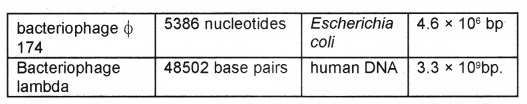
DNA replicates in semi conservative manner was first shown in Escherichia coli by Matthew Meselson and Franklin Stahl performed the following experiment in 1958:
After the completion of replication, each DNA molecule have one Watson-Crick model for parental and one newly synthesised strand. This is termed as semiconservative DNA replication. semiconservative DNA replication.
| They grew E. coli in a medium containing 15NH4Cl. The result was that 15N was incorporated into newly synthesised DNA .This heavy DNA molecule could be distinguished from the normal DNA by centrifugation in a cesium chloride (CsCl) density gradient. |
Then they transferred the cells into a medium with normal 14NH4Cl and took samples at various definite time intervals as the cells multiplied, and extracted the DNA that remained as double-stranded helices.
The DNA that was extracted from the culture one generation after the transfer from 15N to 14N medium [E. coli divides in 20 minutes] had a hybrid DNA.
(Separation of DNA by Centrifugation):
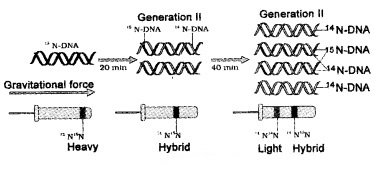
DNA extracted from the culture after another generation [that is after 40 minutes, II generation] was composed of equal amounts of this hybrid DNA and of ‘light’ DNA.
In another experiment radioactive thymidine is incorporated into DNA and was observed the semi-conservative replication of DNA in Vicia faba (faba beans) by Taylor and colleagues in 1958.
2. The Machinery and the Enzymes:
In E. coli, the process of replication takes place with the help of DNA-dependent DNA polymerase, because it uses a DNA template to catalyse the polymerization of deoxynucleotides.
E. coli that has only 4.6 × 106 bp completes the process of replication within 38 minutes. Deoxyribonucleoside triphosphates have double role. Besides acting as substrates, they provide energy for polymerization reaction same as in case of ATP.
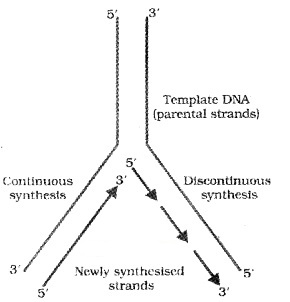
Initially the replication occur within a small opening of the DNA helix (origin of replication) called as replication fork. The DNA- dependent DNA polymerases catalyse polymerization only in one direction, that is 51-3’.
Here on one strand (the template with polarity 3’-5′), the replication is continuous, while on the other (the template with polarity 5′-3′), it is discontinuous. The discontinuously synthesized fragments are later joined by the enzyme DNA ligase.
In eukaryotes, the replication of DNA takes place at S-phase of the cell-cycle. A failure in cell division after DNA replication results into polyploidy(a chromosomal anomaly).
Transcription
The process of copying genetic information from one strand of the DNA into RNA is termed as transcription. In transcription only a segment of DNA is copied into RNA.
Only single stranded RNA is produced by transcription process. If the two RNA molecules are produced simultaneously it would be complementary to each other, hence would form a double stranded RNA. This would prevent the translation.
1. Transcription Unit:
DNA has three regions as transcription unit
- A Promoter
- The Structural gene
- A Terminator
The two strands have opposite polarity and the DNA-dependent RNA polymerase catalyse the polymerisation in only one direction, that is, 5′-3′, the strand that has the polarity 3′-5’acts as a template, and is also referred to as template strand.
The other strand which has the polarity (5′-3′) and the sequence same as RNA (except thymine at the place of uracil), is displaced during transcription. This strand (which does not code for anything) is referred to as coding strand.
For example
| 3′-AT GC ATGC ATGC ATGC ATGC ATGC -5′ Template Strand 5′-TACGTACGTACGTACGTACGTACG-3′ Coding Strand |
The promoter is located towards 5′ – end (upstream) of the structural gene. It provides binding site for RNA polymerase.
The terminator is located towards 3′ – end (downstream) of the coding strand and it usually defines the end of the process of transcription.
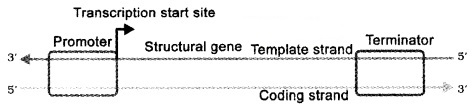
2. Transcription Unit and the Gene:
A gene is the functional unit of inheritance. The DNA sequence coding for tRNA or rRNA molecule also define a gene.
Cistron is a segment of DNA coding for a polypeptide, the structural gene in a transcription unit is called as monocistronic (mostly in eukaryotes) or polycistronic (mostly in bacteria or prokaryotes).
| eukaryotes, the structural genes have interrupted coding sequences – the genes in eukaryotes are split. The coding sequences are exons. The exons are interrupted by introns. |
3. Types of RNA and the process of Transcription:
In bacteria, there are three major types of RNAs:
- mRNA (messenger RNA)
- tRNA (transfer RNA), and
- rRNA (ribosomal RNA).
All three RNAs are needed to synthesise a protein in a cell. The mRNA provides the template, tRNA brings aminoacids and reads the genetic code, and RNAs play structural and catalytic role during translation.
DNA- dependent RNA polymerase that catalyses transcription of all types of RNA in bacteria. RNA polymerase binds to promoter and initiates transcription (Initiation). It uses nucleoside triphosphates as substrate and polymerises in a template and follow the rule of complementarity.
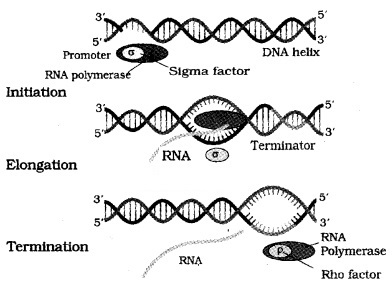
Once the polymerases reaches the terminator region, the nascent RNA and RNA polymerase falls off. This results in termination of transcription.
RNA polymerase catalyse all the three steps, which are initiation, elongation and termination.
The RNA polymerase bind with initiation factor and termination-factor to initiate and terminate the transcription, respectively.
In bacteria, the mRNA does not require any processing and transcription and translation take place in the same compartment.
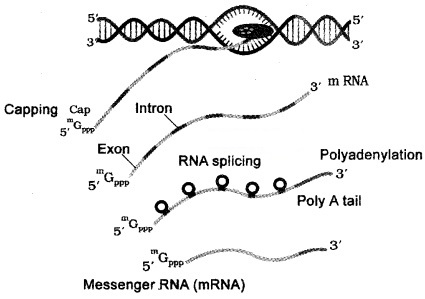
In eukaryotes, there are 3 RNA polymerases in the nucleus. The RNA polymerase I transcribes rRNAs (28S, 18S, and 5.8S).
RNA polymerase III is responsible for transcription of tRNA, 5srRNA, and snRNAs (small nuclear RNAs).
The RNA polymerase II transcribes precursor of mRNA, the heterogeneous nuclear RNA (hnRNA).
| Heterogeneous nuclear RNA contain both the exons and the introns and are non-functional. Hence, it is subjected to a process called splicing where the introns are removed and exons are joined together hnRNA undergo two additional processing called as capping and tailing. In capping an methyl guanosine triphosphate is added to the 5’-end of hnRNA. In tailing, adenylate residues (200-300) are added at 3′-end in a template. It is the fully processed hnRNA, called as mRNA, that is transported out of the nucleus for translation. |
Genetic Code
The process of translation requires transfer of genetic information from a polymer of nucleotides to a polymer of amino acids.
codons for various aminoacids:
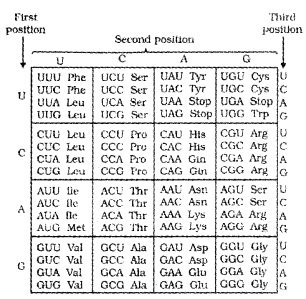
For this, George Gamow, who proposed only 4 bases they have to code for 20 amino acids, the code should constitute a combination of bases.
The code should be made up of three nucleotides( triplet) and in various combination would generate 64 codons, 43 (4 × 4 × 4) Marshall Nirenberg’s cell-free system for protein synthesis helped the code to be deciphered.
Severo Ochoa enzyme- polynucleotide phosphorylase was also helpful in polymerising RNA with defined sequences.
| The salient features of genetic code are: (i) The codon is triplet. 61 codons code for amino acids and 3 codons do not code for any amino acids, hence they function as stop codons. (ii) One codon codes for only one amino acid, hence, it is unambiguous and specific. (iii) Some amino acids are coded by more than one codon, hence the code is degenerate. (iv) The codon is read in mRNA in a contiguous fashion. There are no punctuations. (v) code is nearly universal: for example, from bacteria to human UUU would code for Phenylalanine (phe). Some exceptions to this rule have been found in mitochondrial codons, and in some protozoans. (vi) AUG has dual functions. It codes for Methionine (met), and it also act as initiator codon. |
1. Mutations and Genetic Code:
Deletions and rearrangements in a segment of DNA result in loss or gain of a gene function.
| Example of point mutation is a change of single base pair in the gape for beta globin chain that results in the change of amino acid residue glutamate ‘to valine. It results sickle cell anemia. |
Insertion or deletion of one or two bases, changes the reading frame from the point of insertion or deletion. Such mutations are referred to as frame-shift insertion or deletion mutations.
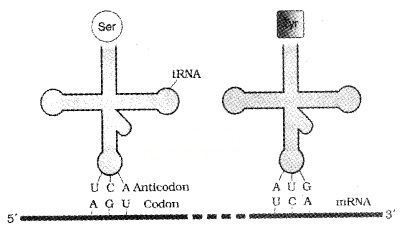
2. tRNA- the Adapter Molecule:
Francis Crick proposed that an adapter molecule would bind to specific amino acids. tRNA has an anticodon loop and an amino acid accepter end to which it binds to amino acids. tRNAs are specific for each amino acid.
For initiation, there is another specific tRNA that is referred to as initiator tRNA. There are no tRNAs for stop codons.
Two-dimensional structure of tRNA looks like a clover-leaf. But in three-dimensional structure of tRNA looks like inverted L.
Translation
Translation is the process of polymerisation of amino acids to form a polypeptide. The order and sequence of amino acids are defined by the sequence of bases in the mRNA. The amino acids are joined by a bond which is known as a peptide bond. Formation of a peptide bond requires energy.
The activation of amino acids with ATP and linked to tRNA- a process commonly called as charging of tRNA or aminoacylation of tRNA. If two such charged tRNAs are brought close together the peptide bond is formed.
The ribosome also acts as a catalyst (23S rRNA in bacteria is the enzyme- ribozyme) for the formation of peptide bond.
A translational unit in mRNA is start codon (AUG) and the stop codon. mRNA also has some additional sequences that are not translated they are called as untranslated regions (UTR). The UTRs are present at both 5′-end (before start codon) and at 3′-end (after stop codon). They are required for efficient translation process.
The ribosome consists two subunits; a large subunit and a small subunit. For initiation, the ribosome binds to the mRNA at the start codon (AUG) that is recognised only by the initiator tRNA.
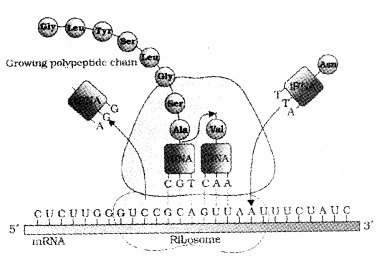
In elongation , amino acid linked to tRNA, and bind to the codon in mRNA by forming complementary base pairs with the tRNA anticodon. The ribosome moves from codon to codon along the mRNA.
Amino acids are added one by one and translated into Polypeptide. At the end, a release factor binds to the stop codon, terminating translation and releasing the complete polypeptide from the ribosome.
Regulation Of Gene Expression
The Lac operon:
In eukaryotes, the regulation is possible in
- transcriptional level (formation of primary transcript),
- processing level (regulation of splicing),
- transport of mRNA from nucleus to the cytoplasm,
- translational level.
For example E. coli synthesised the enzyme beta-galactosidase in the medium if the disaccharide, lactose is present.
Enzyme breakdown the lactose into galactose and glucose; the bacteria use them as a source of energy. The development and differentiation of embryo into adult organisms are also a result of the coordinated regulation of expression of several sets of genes.
In a transcription unit, the activity of RNA polymerase at a promoter is regulated by proteins. These regulatory proteins act both positively (activators) and negatively (repressors). The promoter regions of prokaryotic DNA is regulated by the interaction of adjacent operators. Each operon has its specific operator and specific repressor.
For example, lac operator is present only in the lac operon and it interacts specifically with lac repressor only.
1. The Lac operon:
The function of lac operon was first shown by Jacob and Monod.
| In lac operon the structural gene is regulated by a promoter and regulatory genes. Such arrangement in bacteria is called as operon. |
Other examples are trp operon, ara operon, his operon, val operon, etc.
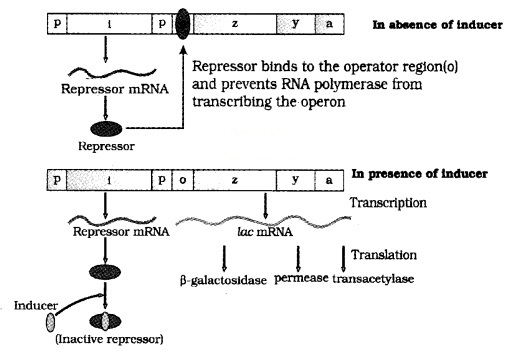
The lac operon consists of one regulatory gene (i gene) and three structural genes (z, y, and a).
| The i gene codes for the repressor of the lac operon. The z gene codes for beta-galactosidase. The y gene codes for permease, which increases permeability of the celt to beta galactosides. The a gene codes for transacetylase. |
Hence, all the three gene products in lac operon are required for metabolism of lactose. Lactose (inducer) is the substrate for the enzyme beta-galactosidase and it regulates switching on and off of the operon.
In the presence of an inducer, such as lactose, the repressor is inactivated by the inducer. Then RNA polymerase bind to the promoter and transcription proceeds. Regulation of lac operon by repressor is referred to as negative regulation.
Human Genome Project
This is mainly aims to find out the complete DNA sequence of human genome If two individuals differ, then their DNA sequences should also be different, at least at some places.
Human Genome Project was launched in the year 1990 (HGP). Human genome consists of approximately 3 × 109 bp.
| Goals of HGP (i) Identify 20,000-25,000 genes in human DNA; (ii) Determine the sequences of the 3 billion chemical base pairs. (iii) Store this information in databases; (iv) Improve tools for data analysis; (v) Transfer related technologies to other sectors, such as industries; (vi) Address the ethical, legal, and social issues (ELSI) that may arise from the project. |
HGP was coordinated by the U.S. Department of Energy and the National Institute of Health. The project was completed in 2003.
This project also aims to solve challenges in health care, agriculture, energy production, environmental remediation.
Methodologies:
For sequencing, the total DNA from a cell is isolated and converted into random fragments of relatively smaller sizes and cloned in suitable host using vectors. The cloning resulted into amplification of each piece of DNA fragment so that it could be sequenced with ease.
The commonly used hosts are bacteria and yeast, and the vectors are called as BAC (bacterial artificial chromosomes), and YAC (yeast artificial chromosomes). The fragments are sequenced using automated DNA sequencers that was developed by Frederick Sanger.
These sequences are then arranged based on some overlapping regions present in them. Alignment of these sequences are done with computer based programs. These sequences are annotated and assigned to each chromosome. The sequence of chromosome 1 was completed only in May 2006.
Another task was the construction of genetic and physical maps on the genpme. This was made possible by knowing the polymorphism of restriction endonuclease recognition sites, and some repetitive DNA sequences.
1. Salient Features of Human Genome:
| (i) The human genome contains 3164.7 million nucleotide bases. (ii) The average gene consists of 3000 bases. The largest known human gene is dystrophin consist of 2.4 million bases. (iii) The total number of genes is estimated at 30,000. The 99.9 per cent nucleotide bases are exactly the same in all people. (iv) The functions are unknown for over 50 per cent of discovered genes. (v) Less than 2 percent of the genome codes for proteins. (vi) Repeated sequences make up very large portion of the human genome. (vii) Repetitive sequences are stretches of DNA sequences that are repeated many times, sometimes hundred to thousand times (viii) Chromosome 1 has most genes (2968), and the Y has the fewest (231) (ix) Scientists have identified about 1.4 million locations where single base DNA differences (SNPs – single nucleotide polymorphism, pronounced as ‘snips’) occur in humans. |
DNA Fingerprinting
The 99.9 per cent of base sequence among humans is the same. The genetic differences between two individuals is calculated by comparing the two sets of 3 × 106 base pairs.
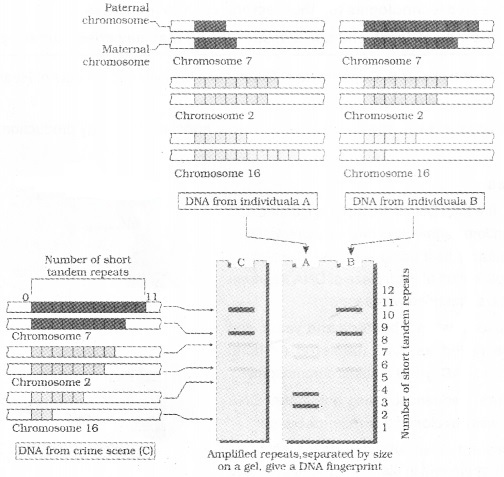
It is the identification of differences in some specific regions in DNA sequence called as repetitive DNA,
These repetitive DNA are separated from bulk genomic DNA as different pegks during density gradient centrifugation.
The bulk DNA forms a major peak and the other small peaks are referred to as satellite DNA. Depending on base composition, length of segment, and number of repetitive units, the satellite DNA is classified into many categories,
- Micro-satellites,
- Mini-satellites etc.
These sequences do not code for any proteins and show high degree of polymorphism. DNA from every tissue (such as blood, hair-follicle, skin, bone, saliva, sperm etc.), of an individual show the same degree of polymorphism, they become very useful identification tool in forensic applications.
The polymorphisms are inheritable from parents to children DNA fingerprinting is the basis of paternity testing, in case of disputes.
The polymorphism in DNA sequence is the basis of genetic mapping and DNA fingerprinting, Polymorphism arises due to mutations. Allelic sequence variation results inheritable mutation.
Such variation are observed in non coding DNA sequence. These mutations accumulating generation after generation, and form one of the basis of variability/polymorphism.
The different types of polymorphisms ranging from single nucleotide change to very large scale changes. For evolution and speciation, such polymorphisms play very important role.
The technique of DNA Fingerprinting was initially developed by Alec Jeffreys. He used a satellite DNA as probe. It is called as Variable Number of Tandem Repeats (VNTR).
The technique, is based on Southern blot hybridisation using radiolabeled VNTR as probe.
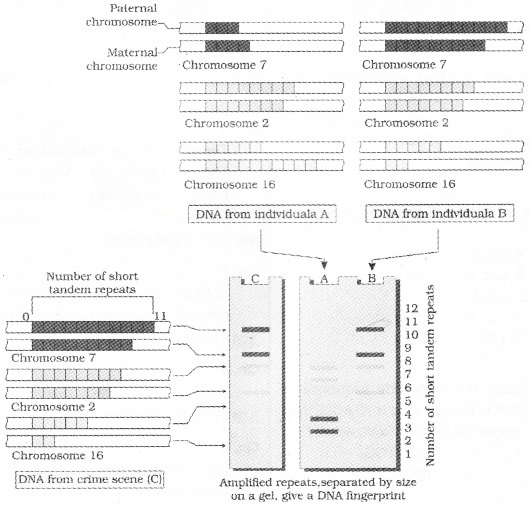
It involves
- isolation of DNA,
- digestion of DNA by restriction endonucleases,
- separation of DNA fragments by electrophoresis,
- transferring (blotting) of separated DNA fragments to synthetic membranes, such as nitrocellulose or nylon,
- hybridisation using labelled VNTR probe, and
- detection of hybridised DNA fragments by autoradiography.
The VNTR belongs mini-satellite.lt is the small DNA sequence. Its copy number varies from chromosome to chromosome in an individual. The numbers of repeat show very high degree of polymorphism.
The size of VNTR varies in size from 0.1 to 20 kb. So after hybridisation with VNTR probe, the autoradiogram gives many bands of differing sizes.
These bands give a characteristic pattern for an individual DNA. It differs from individual to individual in a population except in the case of monozygotic (identical) twins.
We hope the Plus Two Zoology Notes Chapter 4 Molecular Basis of Inheritance help you. If you have any query regarding Plus Two Zoology Notes Chapter 4 Molecular Basis of Inheritance, drop a comment below and we will get back to you at the earliest.
