Plus Two Botany Notes Chapter 4 Biotechnology Principles and Processes is part of Plus Two Botany Notes. Here we have given Plus Two Botany Notes Chapter 4 Biotechnology Principles and Processes.
| Board | SCERT, Kerala |
| Text Book | NCERT Based |
| Class | Plus Two |
| Subject | Botany Notes |
| Chapter | Chapter 4 |
| Chapter Name | Biotechnology Principles and Processes |
| Category | Plus Two Kerala |
Kerala Plus Two Botany Notes Chapter 4 Biotechnology Principles and Processes
Principles Of Biotechnology
The important techniques leads to the origin of modern biotechnology are:
| (i) Genetic engineering: Techniques to alter the chemistry of genetic material (DNA and RNA), and introduce these into host organisms and changing the phenotype of the host organism. (ii) Maintaning the microbial contamination-free condition to promote the growth of desired microbe/eukaryotic cell in large quantities for the manufacture of biotechnological products like antibiotics, vaccines, enzymes, etc. |
In traditional hybridisation the new hybrid formed possess undesirable genes along with the desired genes. But the technique of genetic engineering /recombinant DNA creates transgenic organism contains only the desirable genes.
In a chromosome there is a specific DNA sequence called the origin of replication, which is responsible for the initiation of replication. If the foreign (alien) DNA transferred and integrated into the genome of the recipient, it multiply along with the host DNA. This called as cloning (making multiple identical copies of any template DNA).
| The concept of linking a gene coding for antibiotic resistance with a plasmid of Salmonella typhimurium was the first step in the construction recombinant DNA. It was the work of Stanley Cohen and Herbert Boyer (1972). |
| Antibiotic resistant gene was isolated from a plasmid by cutting DNA at specific locations by restriction enzymes. Then the cut piece of DNA is linked with the plasmid DNA (vectors) with the help of enzyme DNA ligase. |
In the transfer of the malarial parasite into human body, mosquito acts vector. In the same way, a plasmid can be used as vector to deliver an alien piece of DNA into the host organism. It results in the creation of new combination of circular autonomously replicating DNA, it is known as recombinant DNA.
When this DNA is introduced into Escherichia coli, it could replicate using the new host’s DNA and polymerase enzyme to make multiple copies. The ability of forming multiple copies of antibiotic resistance gene in E.coliis called cloning.
The three basic steps in the creation of GMO are
- Identification of DNA with desirable genes;
- Introduction of the identified DNA into the host;
- Maintenance of introduced DNA in the host and transfer of the DNA to its progeny.
Tools Of Recombinant Dna Technology
Important tools are
- restriction enzymes
- polymerase enzymes
- ligases
- vectors and the
- host organism.
1. Restriction Enzymes:
The enzymes restricting the growth of bacteriophage in Escherichia coli is used to cut DNA. This is called restriction endonuclease.
Restriction endonuclease- Hind II cut DNA molecules at a particular point by recognising a specific sequence of six base pairs. This is called recognition sequence.
Another restriction endonuclease EcoRI comes from Escherichia coli RY13. In EcoRI, ‘R’ indicates the order in which the enzymes were isolated from that strain of bacteria.
Restriction enzymes belong to nucleases. These are of two kinds; exonucleases and endonucleases. Exonucleases remove nucleotides from the ends of the DNA whereas, endonucleases bind to the DNA and cut each of the two strands of the double helix at specific points in their sugar-phosphate backbones.
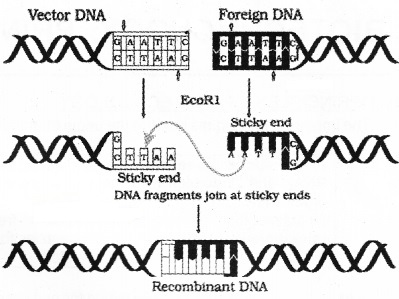
Each restriction endonuclease recognises a specific palindromic nucleotide sequence in the DNA. Actually palindromic nucleotide sequences is the same as the word, “MALAYALAM,” read in both forward and backward.
Eg- 5′ ——GAATTC ——3′
3′ —— CTTAAG —— 5
After cutting DNA duplex by an enzyme, it leaves single stranded portions at the ends. These are sticky ends on each strand. This stickiness of the ends facilitates the action of the enzyme DNA ligase.
Separation and isolation of DNA fragments:
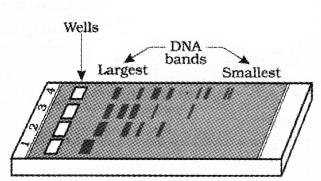
The cut fragments of DNA separated by a technique known as gel electrophoresis. Negatively charged DNA fragments are separated by forcing them to move towards the anode under an electric field through a agarose matrix.
The DNA fragments separate according to their size in agarose gel. So the smaller the fragment size moves farther.
The separated DNA fragments can be visualised only after staining the DNA with ethidium bromide followed by exposure to UV light. It appears as bright orange coloured bands. The separated bands of DNA are cut out from the agarose gel and extracted from the gel piece.
This step is known as elution. The DNA fragments thus obtained are used in constructing recombinant DNA by joining them with cloning vectors.
Agarose gel electrophoresis showing migration of digested and un digested DNA
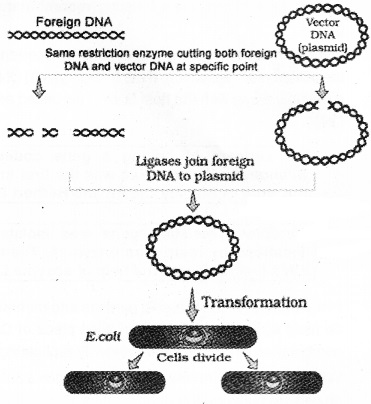
2. Cloning Vectors:
Plasmids and bacteriophages can replicate within bacterial cells independently without chromosomal DNA. Bacteriophages form high copy numbers of their genome within the bacterial cells. Plasmids form 15-100 copies per cell.
If linking an alien piece of DNA with bacteriophage or plasmid DNA, it can multiply its numbers equal to the copy number of the plasmid or bacteriophage. So, this is helpful in the selection of recombinants from non-recombinants.
The features of artificial cloning vector are
(i) Origin of replication (oril):
It is a sequence of cloning vector in which replication starts when any piece of DNA linked to it. This sequence is responsible for controlling the copy number of the linked DNA.
(ii) Selectable marker:
In addition to ‘ori’, the vector contains selectable marker, which helps in identifying and eliminating non transformants and selectively permitting the growth of the transformants.
The genes coding for antibiotic resistance such as ampicillin, chloramphenicol, tetracycline or kanamycin, etc., are considered as selectable markers for E. coli. The normal E. coli cells do not show the resistance against any of these antibiotics.
(iii) Cloning sites:
For linking the alien DNA into the vector, there must be preferably single recognition sites for the commonly used restriction enzymes because more than one recognition sites within the vector results several fragments.
The ligation of alien DNA is carried out at a restriction site present in one of the two antibiotic resistance genes.
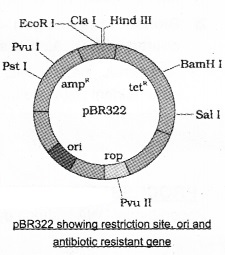
For example, ligation of a foreign DNA at the Bam H I site of tetracycline resistance gene in the vector pBR322, the recombinant plasmids lose tetracycline resistance due to insertion of foreign DNA.
Recombinants selected from non-recombinant by plating the transformants on ampicillin containing medium. The transformants growing on ampicillin containing medium are then transferred on a medium containing tetracycline.
It could not grow in the medium containing tetracycline. But, non-recombinants can grow on both medium Therefore antibiotic resistance gene helps in selecting the transformants.
| Another method of selecting recombinants from non-recombinants is their ability to produce colour in the presence of a chromogenic substrate. For this recombinant DNA is inserted within the coding sequence of an enzyme, beta-galactosidase. This results into inactivation of the enzyme, called as insertional inactivation. |
If the bacteria does not have an insert, chromogenic substrate present in the medium react with betagalactosidase enzyme gives blue coloured colonies.
If the plasmid have an insert, they do not produce any colour due to insertional inactivation of the gene coding for beta galactosidase, these are identified as recombinant colonies.
(iv) Vectors for cloning genes in plants and animals:
Normally Agrobacterioum tumifaciens (a pathogen of several dicot plants) transfer its ‘T-DNA’to normal plant cells and causes tumor. Similarly retroviruses in animals have the ability to transform normal cells into cancerous cells and they are used as vectors for delivering genes of interest to humans.
For delivering genes of interest to plants tumor inducing (Ti) plasmid of Agrobacterium tumifaciens is modified (disarming) as non pathogenic Similarly the retroviruses are disarmed and used to deliver desirable genes into animal cells.
3. Competent Host
(For Transformation with Recombinant DNA)
DNA is a hydrophilic molecule, it cannot pass through cell membranes. For this, bacterial cells must have to be competent to take up DNA.
| This is done by treating them with calcium ions and incubating the cells and recombinant DNA on ice, followed by placing them at 42°C Then putting them back on ice. This helps the bacteria to take up the recombinant DNA. |
Another methods:
- Micro-injection – recombinant DNA is directly injected into the nucleus of an animal cell.
- Biolistics or gene gun -cells are bombarded with high velocity micro-particles of gold or tungsten coated with DNA. It is suitable for plants.
And the last method uses ‘disarmed pathogen’ vectors, which when allowed to infect the cell, transfer the recombinant DNA into the host.
Processes Of Recombinant Dna Technology
Recombinant DNA technology involves several steps. They are
1. Isolation of the Genetic Material (DNA):
Initially the bacterial cells/plant or animal tissue are treated with enzymes such as lysozyme (bacteria), cellulase (plant cells), chitinase (fungus) to open the cell to release DNA along with other macromolecules such as RNA, proteins, polysaccharides, and Other molecules can be removed by appropriate treatments and purified DNA precipitates out afterthe addition of chilled ethanol. It can be observed as collection also lipids.
To get DNA in a pure form and free from other macro-molecules it is treated with enzymes. RNA can be removed by treating with ribonuclease whereas proteins can be removed by treating with protease, of fine threads in the Suspension.
2. Cutting of DNA at Specific Locations:
It is done by incubating purified DNA molecules with the restriction enzyme. Here Agarose gel electrophoresis is used to check the progression of a restriction enzyme digestion. DNA is a negatively charged molecule, hence it moves towards the positive electrode (anode).
After having cut at the source DNA as well as the vector DNA with a specific restriction enzyme, the cut out ‘gene of interest’ from the source DNA and the cut vector with space are mixed and ligase is added. This results in the preparation of recombinant DNA.
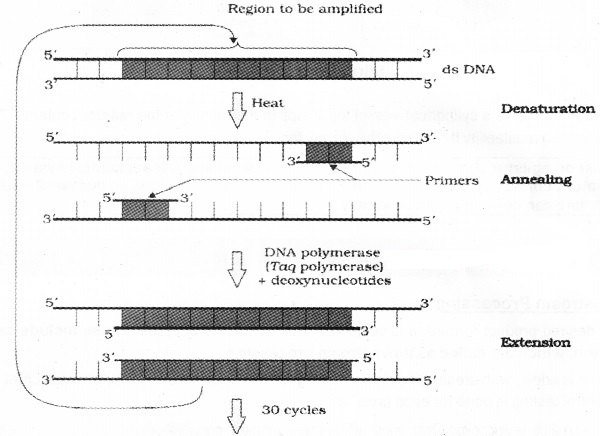
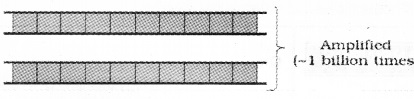
3. Amplification of Gene of Interest using PCR:
| Polymerase Chain Reaction is helpful to produce multiple copies ( eg-1 billion copies-) of the gene of interest. |
It is synthesised in vitro using two sets of primers (chemically synthesised oligonucleotides that are complementary to the regions of DNA) and the enzyme thermostable DNA polymerase (isolated from a bacterium, Thermus aquaticus).
This enzyme extends the primers using the nucleotides provided in the reaction mixture and the genomic DNA as template. If the process of replication of DNA is repeated many times, the segment of DNA (gene of interest) can be amplified. The amplified fragment is used to ligate with vector for further cloning.
(PCR showing denaturation. annealing and extention)
4. Insertion of Recombinant DNA into the Host Cell/Orqanism:
Recombinant DNA carry gene resistant to antibiotic (e.g., ampicillin) is transferred into E. coli cells, the host cells become transformed into ampicillin-resistant cells. If spreading the transformed cells on agar plates containing ampicillin, only the transformants grow and untransformed cells die.
So it is helpful to select a transformed cell in the presence of ampicillin. The ampicillin resistance gene in this case is called a selectable marker.
5. Obtaining the Foreign Gene Product:
The main aim of all recombinant technologies is to produce a desirable protein. Here the foreign gene is expressed under appropriate conditions. If it is necessary to produce target protein i.e recombinant protein on a small scale, rDNA transferred into the host and cloned genes of interest must be grown in the laboratory. Then the protein is extracted and purified.
Stirred tank Bioreactor
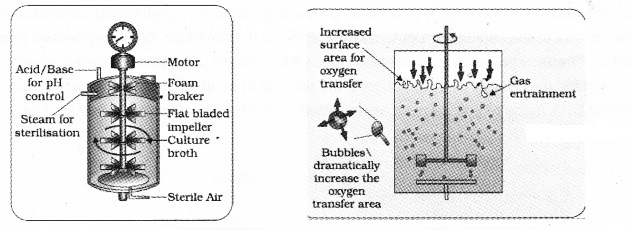
A stirred-tank reactor is a cylindrical vessel that helps in the mixing of the reactor contents. The stirrer also facilitates oxygen availability throughout the bioreactor.
| It consist of agitator system, an oxygen delivery system and a foam control system, a temperature control system, pH control system and sampling ports, so that small volumes of the culture can be withdrawn periodically. |
6. Downstream Processing:
After the desired product formed, it is subjected to a series of processes. These include separation and purification, which are called as downstream processing.
The product is added with preservatives and undergoes clinical trials as in case of drugs. Later the strict quality control testing is done for each product.
We hope the Plus Two Botany Notes Chapter 4 Biotechnology Principles and Processes help you. If you have any query regarding Plus Two Botany Notes Chapter 4 Biotechnology Principles and Processes, drop a comment below and we will get back to you at the earliest.
