ISC Biotechnology Previous Year Question Paper 2019 Solved for Class 12
Maximum Marks: 80
Time allowed: Three hours
- Candidates are allowed additional 15 minutes for only reading the paper. They must NOT start writing during this time.
- Answer Question 1 (Compulsory) from Part I and five questions from Part II, choosing two questions from Section A, two questions from Section B and one question from either Section A or Section B.
- The intended marks for questions or parts of questions are given in brackets [ ].
- Transactions should be recorded in the answer book.
- All calculations should be shown clearly.
- All working, including rough work, should be done on the same page as, and adjacent to the rest of the answer.
Part -1 (20 Marks)
(Answer all questions)
Question 1.
(a) Mention any one significant difference between each of the following : [5]
(i) Anti codon and codon
(ii) Intrinsic fluorescence and extrinsic fluorescence
(iii) Introns and Exons
(iv) Genomic DNA library and cDNA library
(v) RAM and ROM
(b) Answer the following questions: [5]
(i) Which amino acid is optically inactive and why ?
(ii) What is meant by exponential phase 9
(iii) What are designer oils ?
(iv) What is palindromic sequence 9
(v) Which substance is used in diploidization of haploid plants ?
(c) Write the full form of each of the following : [5]
(i) NBRI
(ii) NBTB
(iii) BLAST
(iv) PIR
(v) YAC
(d) Explain briefly the following terms : [5]
(i) Callus
(ii) SNPs
(iii) Lyophilisation
(iv) Gene cloning
(v) Cybrids
Answers:
(a) (i) Anti codon is the corresponding triplet sequence on the transfer RNA (tRNA) which brings in the specific amino acid to the ribosome during translation. The anti codon is complementary to the codon, that is, if the codon is AUU, then the anti codon is UAA.
Codon is the triplet sequence in the messenger RNA (mRNA) transcript which specifies a corresponding amino acid (or a start or stop command).
(ii)
| Intrinsic fluorescence | Extrinsic fluorescence |
| The fluorescence shown by natural compounds is known as intrinsic fluorescence. | There are certain chemical compounds which do not fluorescence. It can be detected after coupling them with a fluorescence probe or flour. This phenomenon is called extrinsic fluorescence. |
(iii)
| Introns | Exons |
| Introns are non-coding DNA base sequences, which are found between exons, but are not transcribed part of mature mRNA. | Exons are coding DNA base sequences that are transcribed into w/RNA and finally code for amino acids in the proteins. |
(iv)
| Genomic DNA Library | cDNA Library |
| (1) It is a collection of clones that represent the complete genome of an organism. (2) It is the mixture of genomic DNA. | (1) The library made from complementary or copy DNA (cDNA) is called cDNA library. (2) cDNA library consists of cDNA clones prepared by using mRNA. |
(v)
| RAM | ROM |
| (1) It stores data and programme temporarily. (2) It contains the data of operating system, software’s and other application programmes. | (1) It provides a non-volatile storage of data. (2) It loads operating system into the memory and starts the computer when switched on. |
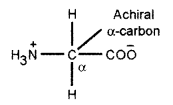
(b) (i) Glycine is the only amino acid which is optically inactive. It is the first neutral amino acid. In Glycine, the central atom has 2 hydrogen atoms. It does not contain any chiral carbon that’s why it is optically inactive.
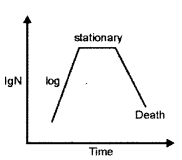
(ii) Exponential Phase : It is a growth phase. In the exponential (log) phase, cells divide as fast as possible according to the growth medium, the microorganism itself and environmental conditions. This phase has a limited duration.
(iii) Designer Oil : “Designer oil” that reduces LDL ( bad ) blood cholesterol levels in humans and increases energy expenditure, which may prevent people from gaining weight. The oil incorporates a phytosterol-based functional food ingredient Phytrol (TM) from Forbes into oil using proprietary technology.
(iv) Palindromic sequence are the base pair sequences that are the same when read forward (left to right) or backward (right to left) from a central axis of symmetry. The sequences read the same on the two strands in 5 → 3 direction and this is also true when we read in the 3 → 5 directions.
(v) Colchicine is used in diploidization of haploid plants. There are mainly two approaches for diploidization-colchicine treatment and endometriosis.
(c) NBRI: National Botanical Research Institute
NBTB : National Biotechnology Board
BLAST: Basic Local Alignment Search Tool
PIR : Protein Information Resource
YAC : Yeast Artificial Chromosome
(d) (i) Callus : It is a mass of meristematic undifferentiated unorganized cells produced in a culture.
(ii) SNPs : Single Nucleotide Polymorphisms (SNPs) are the variations in a nucleotide at genomic DNA in different individuals at a population which occur due to change even in a single base (e.g., A, G, T or C). In human genome, SNPs occur at 1 n – 3.2 million sites.
(iii) Lyophilisation : In this method the microbial suspension is put in small ampoules and frozen through drying under vacuum. Vaccum drying results in sublimation of cell water. The lyophilised cultures are stored at 4°C. The culture remain viable for about 10 years. By this method a large number of cultures are maintained without variation in the characteristics.
(iv) Gene Cloning : It is a process of copying a specific desired gene/section on DNA and producing in large number/quantities.
(v) Cybrids : A cytoplasmicallv hybrid cell with organelles from both parental sources (obtained through fusion of cytoplast with a whole cell) and a nucleus of only one cell Nucleus of the other cell denatured.
Part – II (50 Marks)
(Answer any Jive questions)
Question 2.
(a) With reference to composition of culture medium, answer the following : [4]
(i) Cytokinins
(ii) Auxin’s
(b) Explain the induced fit hypothesis of enzyme action with the help of suitable illustrations. [4]
(c) Write a note on Quaternary’ structure of proteins. [2]
Answers:
(a) (i) Cytokinins:
- It stimulates the cell division process.
- It helps in the morphogenesis of plant cell, along with auxins.
(ii) Auxin:
- It helps in the formation of callus and in the development of xylem along with the promotion of cambial activity.
- It is used for the elongation and enlargement of plant cell and it inhibits the promotion of growths of apical and lateral buds.
(b) The Induced-fit theory:
The key-lock hypothesis does not fully account for enzymatic action; i.e.. certain properties of enzymes cannot be accounted for by the simple relationship between enzyme and substrate proposed by the key-lock hypothesis. A theory called the induced-fit theory retains the key lock idea of a fit of the substrate at the active site but postulates in addition that the substrate must do more than simply fit into the already preformed shape of an active site.
Rather, the theory 7 states, the binding of the substrate to the enzyme must cause a change in the shape of the enzyme that results in the proper alignment of the catalytic groups on its surface. This concept has been likened to the fit of a hand in a glove, the hand (substrate) inducing a change in the shape of the glove (enzyme). Although some enzymes appear to function according to the older key-lock hypothesis, most apparently function according to the induced-fit theory .
Typically, the substrate approaches the enzyme surface and induces a change in its shape that results in the correct alignment of the catalytic groups. In the case of the digestive enzyme carboxypeptidase.
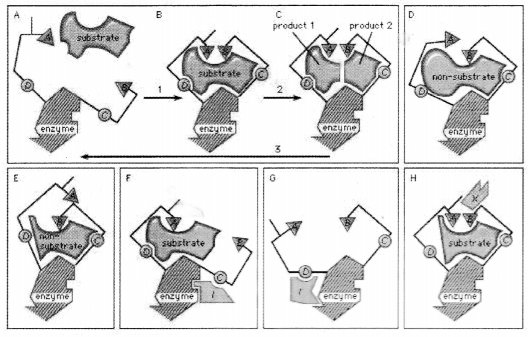
The induced-fit theory 7 explains a number of anomalous properties of enzymes. An example is “non-competitive inhibition.” in which a compound inhibits the reaction of an enzyme but does not prevent the binding of the substrate. In this case, the inhibitor compound attracts the binding group so that the catalytic group is too far away from the substrate to react. The site at which the inhibitor binds to the enzyme is not the active site and is called an allosteric site. The inhibitor changes the shape of the active site to prevent catalysis without preventing binding of the substrate. An inhibitor also can distort the active site by affecting the essential binding group: as a result, the enzyme can no longer attract the substrate.
(c) Quaternary Structure of proteins (4° Structure) : It is the last or fourth level of protein organisation found in only oligomeric proteins or multimers. The multimeric proteins are formed of two to several poly peptides. The monomers or polypeptide sub units are also called protomers. Protomers maybe similar, e.g., two similar a polypeptides in enzyme phosphorvlase. It is known as homogeneous Quaternary structure. An oligomeric protein having dissimilar sub units shows heterogeneous Quaternary structure, .e.g., tetrameric hemoglobin with two ot (141 amino acids each) and two p (146 amino acids each) polypeptide chains.
Question 3.
(a) Explain the important postulates of central dogma. [4]
(b) Name and explain the method used to sterilize the following : [4]
(i) Vitamins
(ii) Forceps and Scalpels
(iii) Nutrient Media
(iv) Explant
(c) What is the Chargaff’s rule of equivalence ? [2]
Answers:
(a) The ‘Central Dogma’ is the process by which the instructions in DNA are converted into a functional product. It was first proposed in 1958 by Francis Crick, discoverer of the structure of DNA.
1. The central dogma of molecular biology explains the flow of genetic information, from DNA to RNA, to make a functional product, a protein.
2. The central dogma suggests that DNA contains the information needed to make all of our proteins, and that RNA is a messenger that carries this information to the ribosomes .
3. The ribosomes serve as factories in the cell where the information is ‘translated’ from a code into the functional product.
4. The process by which the DNA instructions are converted into the functional product is called gene expression.
5. Gene expression has two key stages – transcription and translation .
6. In transcription, the information in the DNA of every cell is converted into small, portable RNA messages.
7. During translation, these messages travel from where the DNA is in the cell nucleus to the ribosomes where they are ‘read’ to make specific proteins.
8. The central dogma states that the pattern of information that occurs most frequently in our cells is :
- From existing DNA to make new DNA (DNA replication)
- From DNA to make new RNA (transcription)
- From RNA to make new proteins (translation)
9. Reverse transcription is the transfer of information from RNA to make new DNA. this occurs in the case of retroviruses, such as HIV . It is the process by which the genetic information from RNA is assembled into new DNA.
Does the ‘Central Dogma’ always apply?
- With modem research it is becoming clear that some aspects of the central dogma are not entirely accurate.
- Current research is focusing on investigating the function of non-coding RNA.
- Although this does not follow the central dogma it still has a functional role in the cell.
(b) Sterilization methods:
(i) Vitamins : Vitamins can be sterilize by the method of autoclaving. For this a concentrated stock solution is required to be prepared followed by fitration and subsequent addition in sterile medium at the temperature of growth (or room temperature).
(ii) Forceps & Scalpels : The metallic instruments like forceps, scalpels, needles, spatulas can be sterilized by flame sterilization method. For this we have to dip them in 25% ethanol ‘ followed by flaming and cooling. It is called incineration.
(iii) Nutrient media : Culture media are properly dispensed in glass container, plugged with cotton or sealed with plastic closures and sterilized by autoclaving (steam sterilization) at 15 psi for 30 minutes. Minimum temperature required for autoclaving of nutrient media is 15 minutes.
(iv) Explant: The explants are sterilized by disinfectants (e g., sodium, hypochlorite. NaOCl, mercuric chloride-HgCl2) and washed aseptically for 6-10 times with sterlized distilled water.
(c) Chargaff’s rule of equivalence : Deoxy ribonucleic Acid (DNA), the genetic material is made up of four types of organic nitrogenous bases : adenine (A), guanine (G), thymine (T) and cytosine (C). Of these, A and G are the purines and T and C are the pyrimidines. Chargaff gave the base pairing rule or the rule of base equivalence which states that only one purine can combine with one pyrimidine. That means A can combine with T and G with C. Two purines or two pyrimidines cannot combine with each other; if they do so, there w ill be a sudden change in the characteristic of an organism. This sudden change is called mutation.
Question 4,
(a) Differentiate between oils and fats. Discuss hydrolysis, rancidity and hardening shown by lipids. [4]
(b) Using tissue culture method one can produce disease free plants. Discuss the method used to produce virus free plants. [4]
(c) Write the main objectives of HGP. [2]
Answer:
(a) Difference between Fats and Oils :
| Fats | Oils |
| 1. Solid at room temperature | 1. Liquid at room temperature |
| 2. Saturated and trans are its types | 2. Unsaturated fats like monounsaturated and polyunsaturated are its types |
| 3. Mostly derived from animal | 3. Mostly derived from plants |
| 4. Increases cholesterol levels | 4. Improves cholesterol levels |
| 5. Mainly comes from animal food but also through vegetable oil by process called hydrogenation Example: Butter, beef fat Contains 9 cal/gm | 5. Mainly comes from plants or fish Example:Vegetable oil, fish oil Contains 9 cal/gm |
Hydrolysis of lipids: Hydrolysis is the breakdown of a substance by the addition of water. Fats and oils are hydrolyzed by moisture to yield glycerol and 3 fatty acids. This leads to hydrolytic rancidity of food product characterized by unpleasant flavor and aroma thereby making it undesirable for consumers. Chemically fats are esters, so they are liable to hydrolysis. This reaction is catalyzed by lipase enzyme or can occur via non-enzymatic hydrolysis. Partial hydrolysis of triglycerides will yield mono- and di- glycerides and free fatty’ acids.
Rancidity: Rancidity is the natural process of decomposition of fats or oils by either hydrolysis or oxidation or by both. The process of degradation converts fatty acids esters of oils into free fatty acids. This gives rise to an unpleasant odour and taste in food. These lipids degrade to the point of becoming either unpalatable or unhealthy to ingest.
Hydrogenation and hardening: Hydrogenation of unsaturated fatty acids is widely practiced. This treatment affords saturated fatty acids. The extent of hydrogenation is indicated by the iodine number. Hydrogenated fatty- acids are less prone toward rancidification. Since the saturated fatty acids are higher melting than the unsaturated precursors, the process is called hardening.
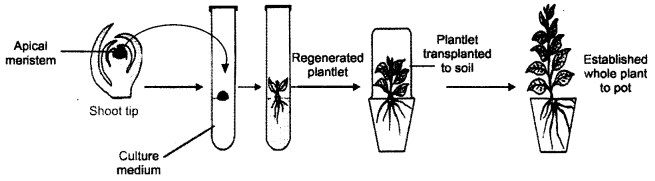
(b) Production of virus-free plants : Plant viruses are found in nature and propagate inside the cells of living plant. There are many plants that suffer from serious diseases developing a variety of symptoms such as mosaic, chlorosis, necrosis, vein clearing, lead curling, leaf rolling, etc. Sometimes the disease severity is so high that viruses migrate to vascular bundles and also get established inside the seeds.
For the first time G Moral and C. Martin (1952) produced virus-free shoots of Dahlia from virus-infected plants through shoot meristem culture, On 26th November, 2002 a US Patent was granted to H.C. Chaturvedi and co-workers for successfully regenerating and proliferating shoot meristem of Cirus spp.
Using tissue culture method we can produce disease-free plants. Generally, the apical meristem of plants are free from viruses and the other microorganisms. However, if viral particles are present, their number would be quite low.
Methods of virus elimination :
There are two methods that are used to produce virus-free plants.
(i) Heat Treatment of Meristem : Before meristem culture, viruses associated with meristem are eliminated in vitro by heat treatment (thermotherapy) of whole plants. Heat treatment is given through hot water or air at 35-40°C for a varying period (a few minutes to several months). It should be noted that prolonged heat treatment inactivates that host resistance. Percentage of plant survival is also low. Temperature tolerant viruses may survive inside the plant tissue.
(ii) Meristem-tip Culture : Meristem-tip culture is the most widely applicable approach for virus elimination. In most of the cases explant (shoot tip) of 100-1000 pm has been cultured to raise virus-free plants. But the explant of small size (1 mm) i.e., meristem tip is preferred for in-vitro culture. Meristem tip is excised in aseptic conditions and cultured on nutrient medium. If required thermotherapy is given to the mother plant before excision of meristem tip. The inoculated tubes are incubated properly in light and dark regime (Fig.)
There are some chemicals which are used to eliminate viruses from plant tissues and protoplasts. These are malachite green, thiouracil, acetyl salicylic acid, cycloheximide, actinomvcin-D, etc.
(c) Objectives of HGP :
(1) The goals of the original HGP were not only determine all 3 billion base pairs in the human genome with a minimal error rate, but also to identify’ all the genes in this vast amount of data. This part of the project is still ongoing although a preliminary count indicates about 30,000 genes in the human genome, which is far fewer than predicated by most scientists.
(2) Another goal of the HGP was to develop faster and more efficient methods for DNA sequencing and sequence analysis and the transfer of these technologies to industry.
(3) Another goal of the HGP is the study of its ethical, legal, and social implications. It is important to research these issues and find the most appropriate solutions before they become large dilemmas. Otherwise its effect will manifest in the form of major political concerns.
Question 5.
(a) Discuss the mechanism of the lac operon model of regulation of gene expression. [4]
(b) Give four points of difference between southern blotting technique and northern blotting technique. [4]
(c) Give four characteristics of genetic code. [2]
Answers:
(a) The lac Operon Model: The lac operon model is an example of transcriptional regulation of gene expression. The lac operon consists of a promoter, an operator and three structural genes (Fig.). RNA polymerase binds to promoter site of DNA. This result in start of sequential transcription of structural genes (lac Z, lac Y and lac A). The lac Z expresses gene β-galactosidase, lac Y encodes permease and lac A synthesizes transacetylase. The operator site is located between the promoter and first structural gene (lac Z). The repressor protein, expressed by the regulator gene, binds to operator. The structural gene (lac Z. lac Y and lac A) do not express enzymes (A).
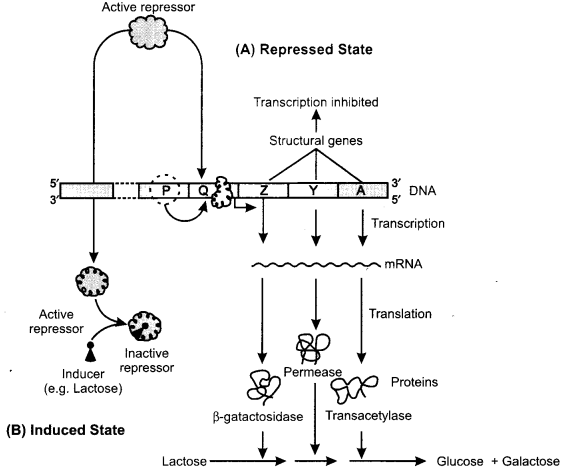
The repressor protein expressed by the repressor gene (which is not a part of operon) involves the transcription of structural genes. Transcription of structural genes is stopped when repressor protein is attached on the operator gene. If lactose is supplied, it enters the cell and binds with repressor protein. Then repressor protein is inactivated due to change in conformation of the repressor protein. Therefore, repressor fails to attach on the operator gene, and the structural genes are transcribed into an wRNA which in turn translates the proteins (B).
Here, lactose acts as anti-inducer but not a real inducer. Allolactose (an inducer of lactose) is a natural inducer for the lac operon. In regulation of lac operon, lactose is taken up. Then it is broken by p-galactosidase into glucose and galactose. Galactose is converted into allolactose and lactobiose through a side reaction with lactose. The allolactose induces the operon. Consequently, lactose is utilized enzymatically.
(b) Southern Blotting technique involves separation and identification of a specific DNA fragment. In Northern Blotting technique, the RNA is analysed rather than DNA. During southern blotting NaOH denatures DNA to form single stranded DNA which are transferred from gel to nitrocellulose membrane. In northern blotting, total RNA molecules are extracted and then mRNA molecules are isolated by using oligo (dT) cellulose Chromatography. RNA samples separated are transferred to a nylon membrane.
(c) Characteristics of Genetic Code :
(i) Triplet code : Three adjacent nitrogen bases constitute a codon which specifies the placement of one amino acid in a polypeptide.
(ii) Start signal: Polypeptide synthesis is signaled by AUG or methionine codon and GUG — Valine codon. They have dual function.
(iii) Stop signal: Polypeptide chain termination is signaled by three termination codons — UAA, UAQ and UGA. They do not specify any amino acid and are hence also called non-sense codon.
(iv) Universal code : The genetic code is applicable universally i.e. the codon specifies the same amino acid from a virus to a tree or human being.
(v) Non-ambiguous codon : One codon specifies only one amino acid and not any other.
Question 6.
(a) With reference to vector less methods of gene transfer explain each of the following : [4]
(i) Liposome mediated gene transfer
(ii) Electroporation
(iii) Transfection
(iv) Transformation
(b) With reference to application of tissue culture techniques, explain the following : [4]
(i) Haploid production
(ii) Triploid production
(c) What is meant by DNA probe ? [2]
Answers:
(a) The term direct or vector less transfer of DNA is used when the foreign DNA is directly introduced into the plant genome. Direct DNA transfer methods rely on the delivery of naked DNA into the plant cells. This is in contrast to the Agrobacterium or vector-mediated DNA transfer which may be regarded as indirect methods. Majority of the direct DNA transfer methods are simple and effective. And in fact, several transgenic plants have been developed by this approach.
1. Liposome-Mediated Transformation: Liposomes are artificially created lipid vesicles containing a phospholipid membrane. They are successfully used in mammalian cells for the delivery of proteins, drugs etc. Liposomes carrying genes can be employed to fuse with protoplasts and transfer the genes.The efficiency of transformation increases when the process is carried out in conjunction with polyethylene glycol (PEG). Liposome-mediated transformation involves adhesion of liposomes to the protoplast surface, its fusion at the site of attachment and release of plasmids inside the cell.
2. Electroporation: Electroporation basically involves the use of high field strength electrical impulses to reversibly permeabilize the cell membranes for the uptake of DNA. This technique can be used for the delivery of DNA into intact plant cells and protoplasts. The plant material is incubated in a buffer solution containing the desired foreign/target DNA, and subjected to high voltage electrical impulses. This results in the formation of pores in the plasma membrane through which DNA enters and gets integrated into the host cell genome.
3. Transfection: Transfection is the process of inserting genetic material, such as DNA and double stranded RNA, into mammalian cells. The insertion of DNA into a cell enables the expression, or production of proteins using the cell’s own machinery, whereas insertion of RNA into a cell is used to down-regulate the production of a specific protein by stopping translation. While the site of action for transfected RNA is the cytoplasm, DNA must be transported to the nucleus for effective transfection. Therefore, the DNA can be transiently expressed for a short period of time, or become incorporated into the genomic DNA, where the change is passed on from cell to cell as it divides.
4. Transformation: Transformation is the method of introducing foreign DNA into bacterial cells (e.g. E.coli). The uptake of plasmid DNA by E.coli is carried out in ice-cold CaCl2 (0-5°C), and a subsequent heat shock (37-45°C for about 90 sec). By this technique, the transformation frequency, which refers to the fraction of cell population that can be transferred, is reasonably good e.g. approximately one cell for 1000 (10-3) cells.The mechanism of the transformation process is not fully understood. It is believed that the CaCl2 affects the cell wall, breaks at localized regions, and is also responsible for binding of DNA to cell surface. A brief heat shock (i.e. the sudden increase in temperature from 5°C to 40°C) stimulates DNA uptake. In general, large-sized DNAs are less efficient in transforming.
(b) (i) Applications of triploid plants :
- Autotriploidy induces gigas effect i.e., large-sized leaves, flowers, fruits and seeds.
- Autotriploids are sterile (with faulty pairing and distribution of chromosomes and abnormal gametes) and reproduce vegetatively only. They are useful for the production of seedless plants.
(ii) Applications of haploid plants :
- Haploid plants have been used in breeding programmes of rice, wheat, barley. Brassica spps., tobacco, potato, etc.
- A doubled haploid strain are produced in only two years a net saving of 4 years for the production of homozygous lines by selfing or close breeding of both self and cross pollinated crops. They may be released directly as a variety if it performs well in yield trials.
- Haploid plants are most suitable for the study of recessive mutations.
(c) DNA Probe : It is a solution of radioactive, single-stranded DNA or oligodeoxy nucleotides (a DNA segment of few to several nucleotide’s). The name probe signifies the fact that this DNA molecule is used to detect and identify’ the DNA fragment in the gel/membrane that has a sequence complementary to the probe. The probe hybridizes with the complementary DNA on the membrane to the greater extent with a low non-specific binding on the membrane. This step is known as hybridization reaction.
Question 7.
(a) Explain how biotechnology helps in developing following traits in crops : [4]
(i) Biodegradable plastic
(ii) Pest resistance
(iii) Drought resistance
(iv) Salinity resistance
(b) Write the principle and applications of the following techniques : [4]
(i) Hydrophobic interaction
(ii) Colorimetry
(c) What are start and stop codoms ? [2]
Answers:
(a) (i) Biodegradable plastic : The biodegradable plastic is made from lactic acid which is produced at the time of bacterial fermentation of plant materials like discarded stalks of com. Biodegradable plastic is a material w hich has most of the properties of plastic except the property of being non-biodegradable. The biodegradable plastic polyhydroxyalkanates e.g., poly hydroxyl butyrate (PHB) are obtained commercially by fermentation with bacterium Alcaligenes eutrophus. The genetically engineered Arabidopsis plants produced poly hydroxyl butyrate (PHB) globules in their chloroplasts without effecting plant growth and development. The large scale of poly hydroxybutyrate (PHB) can be extracted from leaves as well as from transgenic plants.
(ii) Pest resistance: To reduce the loss by the way of killing insects, farmers apply insecticides (i.e., synthetic pesticides) on the crop plants. However, the synthetic insecticides pose a serious threat to the health of plants, animals and humans. The alternative and novel ways of rescue from damages of insects are the use of transgenic technology. It is eco-friendly , cost-effective, sustainable and effective way of insect control. Using biotechnological approaches many transgenic crops having cry’ gene i.e., Bi-genes have been developed and commercialized. Some examples of Bt-crops are brinjal, cauliflower, cabbage, canola, com, cotton, egg plant, maize, potato, tobacco, tomato, rice, soyabean, etc.
(iii) Drought resistance : There are some plants that resist drought conditions. The property of resistance against water loss from the surface of plants is naturally checked by the plants. The special feature is controlled by a specific gene called ‘drought resistance (DR) gene’. Plant biotechnology offers the introduction of DR gene from drought resistant plants into the staple crops for the benefit of enhanced crop yield. The promising GM technology includes the use of techniques for increasing drought tolerance. Research works on these aspects are being investigated at the International Maize and Wheat Improvement Center, Mexico and individual countries itself.
(iv) Salinity resistance : Irrigation has enabled the transformation of arid regions into some of the w orld’s most productive agricultural areas. Excess salinity, however, is becoming a major problem for agriculture in dry parts of the world. In several cases, scientists have used biotechnology to develop plants with enhanced tolerance to salty conditions.
Another approach to engineering salt tolerance uses a protein that takes excess sodium and diverts it into a cellular compartment w here it does not harm the cell. In the lab. this strategy was used to create test plants that were able to flow er and produce seeds under extreme salt levels. Commercially available crops with such a modification are still several years away.
(b) (i) Hydrophobic Interaction : Hydrophobic interaction chromatography (HIC) is a classic purification tool applied in protein and antibody, laboratory and industrial production process. It has been mainly used for the removal of both product-related impurities such as aggregates, as well as process contaminants such as host cell proteins. This review will focus on the recent development of HIC in its applications in the industrial purification processes. The process economy and requirements of high product purity and quality hare driven much of the recent advancement in HIC chromatography in terms of increased throughput and enhanced selectivity or resolution. Meanwhile, high throughput screening (HTS).
design of experiments (DoE) and platform approach for process development hare been applied to shorten the development time. The throughput improvement has been achieved through new resins with increased binding capacity , using dual salts for load conditioning, and operating in the flow-through mode. In addition, hydrophobic interaction membrane filter chromatography technology reduces bed volumes and buffer usage and potentially improves process throughput by reducing cycle time. Selectirity and/or resolution enhancements have been achieved through optimization of operation parameters such as temperature and efforts such as application of solvent additives
(ii) Colorimetry: It is based on the use of interaction of light energy with colored solutions of certain molecules as when light passes through a colored solution, some wavelengths are absorbed more than others. The amount of light absorbed is proportional to the intensity of color and hence to the concentration of the compound.
Applications:
- Quantitative estimation i.e., concentration in solution.
- Detect and identification of bio molecules.
(c) START codons: The codonAUGis called the START codon as it the first codon in the transcribed mRNA that undergoes translation. AUG is the most common START codon and it codes for the amino acid methionine (Met) in eukaryotes and formyl methionine (fMet) in prokaryotes. During protein synthesis, the tRNA recognizes the START codon AUG with the help of some initiation factors and starts translation of mRNA.
STOP codons : There are 3 STOP codons in the genetic code – UAG UAA, and UGA. These codons signal the end of the polypeptide chain during translation. These codons are also known as nonsense codons or termination codons as they do not code for an amino acid.
The three STOP codons have been named as amber (UAG), opal or umber (UGA) and ochre (UAA). “Amber” or UAG was discovered by Charles Steinberg and Richard Epstein and they named it amber after the German meaning of the last name of their friend Harris Bernstein. The remaining two STOP codons were then named “ochre” and “opal” so as to maintain the “color names” theme.
Question 8.
(a) List any four responsibilities carried out by NCBI. [4]
(b) Give a comparative account of cell differentiation, dedifferentiation, rediffemtiation and vascular differentiation. [4]
(c) What is the difference between dNTP and ddNTP ? [2]
Answers:
(a) Responsibilities carried out by NCBI:
1. It conducts research on fundamental biomedical problems at the molecular level using mathematical and computational methods.
2. It maintains collaborations with several NIH institutes, academia, industry, and other governmental agencies.
3. It fosters scientific communication by sponsoring meetings, workshops, and lecture series.
4. It supports training on basic and applied research in computational biology for postdoctoral fellows through the NIH Intramural Research Program.
5. It engages members of the international scientific community in informatics research and training through the Scientific Visitors Program.
6. It develops, distributes, supports, and coordinates access to a variety of databases and software for the scientific and medical communities.
7. It develops and promotes standards for databases, data deposition and exchange, and biological nomenclature.
(b) Cell Differentiation : The cells derived from root apical meristem (RAM) and shoot apical meristem (SAM) and cambium differentiate, mature to perform specific functions. This act leading to maturation is termed differentiation. They undergo a few or major structural changes both in their cell walls and protoplasm. For example, to form, a trachery element the cells lose its protoplasm but develops a very strong, elastic, lignocellulosic secondary cell wall is best suited to carry water to long distances even under extreme tension. Similarly, cells designated to be mesophyll come to possess many chloroplasts so as to, perform photosynthesis. On one hand, a parenchyma in hydrophytes develop large schizogenous interspaces for mechanical support, buoyancy and aeration, but on the other hand, in a potato tuber (or perennating organs) develops more amyloplasts.
Dedifferentiation : In plants, the living differentiated cells can regain the capacity to divide mitotically under certain conditions. The sum of events, that bestow this capacity to divide once again, are termed dedifferentiation. A dedifferentiated tissue can act as meristem (e.g., interfascicular vascular cambium, wound meristem, cork cambium).
Redifferentiation: The product of dedifferentiated cells/tissue which lose the ability to divide are called redifferentiate cells/tissues and the event, redifferentiation. However, the growth in plants is open, and even differentiation in plants is open, because, e g., the same apical meristem cells give rise to xylem phloem, fibers, etc., cells/tissues arising out of same meristem have different structures at maturity. The final structure at maturity of a cell/ tissue arising out of same meristem is determined by the location of the cell within.
Vascular Differentiation : Vascular tissues, xylem and phloem, are differentiated from meristematic cells, procambium, and vascular cambium. Auxin and cytokinin have been considered essential for vascular tissue differentiation: this is supported by recent molecular and genetic analyses.
(c) dNTP lacks-OH at 2’ position of ribose while ddNTP lack-OH both at 2 and 3’ position of ribose sugar. Using ddNTP’s during sequencing, chain elongation can be terminated at 3’ position of ribose.
Question 9.
(a) Proteins have many important functions in an organism. Justify the statement giving its various
roles with an example of each. [4]
(b) With reference to screening strategies, explain the following : [4]
(i) Insertional Inactivation method
(ii) Blue – White method
(c) How was insulin obtained before the advent of rDNA technology ? [2]
Answers:
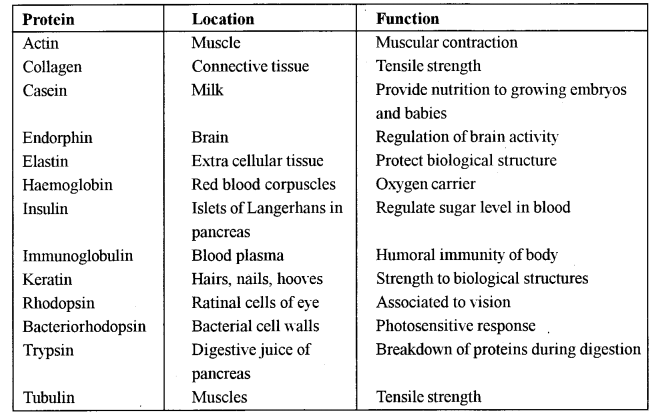
(a) Proteins are the most abundant intracellular macro molecules. These constitute more than half of dry weight of most organisms. Some proteins provide structural elements of the body such as hairs, collagens, enzymes are oxygen carriers.
There are 100,000 kinds of proteins in humans. They are most important for cell functions. Proteins are the linear polymers of 20 amino acids. The amino acids differ from each other in having different R-groups. The R-groups may be changed. They may be polar, non-polar, aromatic or hydrophobic. Amino acid sequences in different proteins differ. This difference is due to diversity in R-groups of 20 amino acids. Their specific location and functions are given in Table. Proteins are grouped into different types on the basis of their biological functions.
b) (i) Insertional inactivation: Harder problem to solve is to determine which of the transformed colonies comprise cells that contain recombinant DNA molecules, and which contain self- ligated vector molecules. Insertional inactivation is the inactivation of a gene by inserting a fragment of DNA into the middle of its coding sequence. Any future products from the inactivated gene will not work because of the extra codes added to it. Recombinants can, therefore, be identified because the characteristic coded by the inactivated gene is no longer visible.
pBR322 contains genes which code for ampicillin resistance and tetracycline resistance, BamH1 cuts in the middle of the gene which codes for tetracycline resistance. If a gene is inserted here, the plasmid loses it ability to code for tetracycline resistance. Thus, the plasmid containing the recombinant gene is resistant to ampicillin but sensitive to tetracycline. To screen, we use replica plates.
(ii) The blue-white screen is a molecular technique that allows for the detection of successful ligation’s in vector-based gene cloning. DNA of interest is ligated into a vector. The vector is then transformed into competent cell (bacteria). In this method, a reporter gene lac Z is inserted in the vector. The lac Z encodes for the enzyme β- galactosidase which breaks a synthetic substrate X-gal (5-bromo-4-chloro-indolyl, β-D-galacto-pyranoside) into insoluble blue colored product. The competent cells are grown in the presence of X-gal. If the ligation was successful, the bacterial colony will be white because β- galactosidase is not synthesized due to the inactivation of lac Z; if not, the colony will be blue. This technique allows for the quick and easy detection of successful ligation, without the need to individually test each colony. An example of such a vector is the artificially reconstructed plasmid pUC19.
(c) Insulin is a hormone that regulates the amount of glucose (sugar) in the blood and is required for the body to function normally. People who do not produce the necessary amount of insulin have diabetes. There are two general types of diabetes. The most severe type, known as Type I or juvenile-onset diabetes, is when the body does not produce any insulin. Type I diabetics usually inject themselves with different types of insulin three to four times daily. Dosage is taken based on the person’s blood glucose reading, taken from a glucose meter. Type II diabetics produce some insulin, but it is either not enough or their cells do not respond normally to insulin.
History : Before researchers discovered how to produce insulin, people who suffered from Type I diabetes had no chance for a healthy life. Then in 1921, Canadian scientists Frederick G Banting and Charles H. Best successfully purified insulin from a dog’s pancreas. Over the years scientists made continual improvements in producing insulin. In 1936. researchers found a way to make insulin with a slower release in the blood. They added a protein found in fish sperm, protamine, which the body breaks down slowly. One injection lasted 36 hours. Another breakthrough came in 1950 when researchers produced a type of insulin that acted slightly faster and does not remain in the bloodstream as long. In the 1970’s, researchers began to try and produce an insulin that more mimicked how the body’s natural insulin worked: releasing a small amount of insulin all day with surges occurring at mealtimes.
Researchers continued to improve insulin but the basic production method remained the same for decades. Insulin was extracted from the pancreas of cattle and pigs and purified. The chemical structure of insulin in these animals is only slightly different than human insulin, which is why it functions so well in the human body. (Although some people had negative immune system or allergic reactions.) Then in the early 1980’s biotechnology revolutionized insulin synthesis. Researchers had already decoded the chemical structure of insulin in the midi 1950’s. They soon determined the exact location of the insulin gene at the top of chromosome 11. By 1977, a research team had spliced a rat insulin gene into a bacterium that then produced insulin.
