ISC Biotechnology Previous Year Question Paper 2014 Solved for Class 12
Maximum Marks: 80
Time allowed: Three hours
- Candidates are allowed additional 15 minutes for only reading the paper. They must NOT start writing during this time.
- Answer Question 1 (Compulsory) from Part I and five questions from Part II, choosing two questions from Section A, two questions from Section B and one question from either Section A or Section B.
- The intended marks for questions or parts of questions are given in brackets [ ].
- Transactions should be recorded in the answer book.
- All calculations should be shown clearly.
- All working, including rough work, should be done on the same page as, and adjacent to the rest of the answer.
Part-I
(Answer all questions)
Question 1.
(a) Mention any one significant difference between each of the following : [5]
(i) RNA polymerases and Tag DNA polymerases
(ii) ln-situ conservation and Ex-situ conservation
(iii) Micronutrients and Macronutrients
(iv) mRNA and tRNA
(v) Essential amino acids and Non-essential amino acids
(b) Answer the following questions : [5]
(i) Why is primer essential during DNA replication ?
(ii) Name a pest resistant crop developed using biotechnology techniques.
(iii) Why are glucose and lactose referred to as reducing sugars ?
(iv) What is the role of dcMTP during DNA sequencing ?
(v) How is the disease alkaptonuria caused ?
(c) Write the Ml form of the following: [5]
(i) BLAST
(ii) ESTS
(iii) MOD
(iv) RAM
(v) SNP
(d) Explain briefly: [5]
(i) Splicing
(ii) Mitotic arrest
(iii) Reverse transcription
(iv) Interferon
(v) Activation energy of enzymes
Answer:
(a) (i)RNA Polymerase: It is an enzyme found in both prokaryotic and eukaryotic cells. It catalyzes the transcription of DNA to synthesize precursors of tmRNA.
Taq DNA Polymerase: It is a thermostable DNA polymerase, originally isolated from bacterium, Thermus aquaticus. It is used in polymerase chain reaction.
(ii) In-situ conservation: It means on-site conservation. It is the process of protecting an endangered plant or animal species in its natural habitat either by protecting or cleaning up the habitat itself, or by defending the species from predators.
Ex-situ conservation: It means literally, “off-site conservation”. It is the process of protecting an endangered species of plant or animal by removing part of the population from a threatened habitat and placing it in a new location, which may be a wild area or within the care of humans e.g., Zoos, Botanical garden, etc.
(iii) Micronutrients: Micronutrients are essential elements required by organisms in small quantities. They include microminerals and vitamins.
Macronutrients: Macronutrients include carbohydrates, proteins and fats required in larger quantities than micronutrients in the body
(iv) mRNA: mRNA is present in nucleus and functions in cytoplasm. It carries messages from DNA.
tRNA: tRNA is an adapter present in cytoplasm. It recognizes and brings amino acids near ribosomes for protein synthesis.
(v) Essential amino acid: The essential amino acids cannot be synthesized by the human body and are obtained from food.
Non-essential amino acid: The non-essential amino acids can be synthesized by the human body. They can be produced from other amino acids and substances in the diet and metabolism.
(b) (i) A primer is a strand of short RNA that functions as a starting point for DNA replication. It is required because the enzymes DNA polymerases that catalyze replication, can only add new nucleotides to an existing strand of DNA.
(ii) Bt crop is a pest resistant crop developed by using biotechnology techniques.
(iii) Glucose and lactose are the reducing sugars as they have an open chain with a free aldehyde or a ketone group.
(iv) ddNTP’s are a form of nucleotide that inhibit extension of the primer in DNA sequencing. Once a ddNTP has been incorporated into the DNA chain, it halts the process.
(v) Alkaptonuria (black mine disease or alcaptonuria): It is a rare, inherited recessive genetic disorder of tyrosine metabolism, caused due to mutation of gene.
Symptoms : Black urine, ochronosis, leading to osteoarthritis, kidney stones.
(c) (i) BLAST : Basic Local Alignment Search Tool
(ii) EST : Expressed Sequence Tag
(iii) MGD : Mouse Genome Database
(iv) RAM : Random Access Memory
(v) SNP : Single Nucleotide Polymorphism.
(d) (i) Splicing is the process by which introns(Non-coding) are removed from heteronuclear RNA to produce mature functional messanger RNA by uniting exons.
(ii) The process by which the mitotic cell cycle is stopped during one of the normal phases G1S, G2,M) of cell cycle.
(iii) It is the process by which DNA is synthesized from RNA template by reverse transcriptase enzyme. This process may be particularly helpful in treating diseases like HIV.
(iv) Interferon is a protein released by animal cells, generally in response to the presence of a virus. It inhibits virus replication.
(v) Activation energy of enzyme is the amount of energy required to bring substrates together to the point where they can react.
Part-II
(Answer any five questions)
Question 2.
(a) What are polysaccharides ? Name any three naturally occurring polysaccharides and give their structural units. [4]
(b) State one important use of each of the following in biotechnology : [4]
(i) Genomic DNA library and cDNA library
(ii) Transfection and Transformation
(c) What are single cell proteins ? [2]
Answer :
(a) Polysaccharides are the complex polymeric carbohydrate molecules composed of long chains of monosaccharide units linked together by glycosidic bonds. They vary in structure from linear to highly branched. Examples include storage polysaccharides such as starch and glycogen, and structural polysaccharides such as cellulose and chitin. Starch, glycogen and cellulose are the natural polysaccharide and their structural units are glucose.
(b) (i) Genomic Library: This is a collection of clones that represent the complete genome of an organism. For construction of a Genomic library the entire genomic DNA is isolated from host cells or tissues, purified and broken randomly into fragments of correct size for cloning into a suitable vector.
The major use of Genomic library is hierarchichal shotgun sequencing.
cDNA Library : The library made from complementary or copy DNA (cDNA) is called cDNA library. The library represents the DNA of only eukaryotic organisms, not the prokaryotic once.
cDNA library’ is used to express eukaryotic gene in prokaryotes. cDNA libraries are most usefirl in reverse genetics where the additional genomic information is of less use. It is also useful for subsequently isolating the gene that codes for that mRNA.
(ii) Transformation : In the Biotechnology, Transformation means introduction of rDNA molecules into a living cell. It is the method of transfer of recombinants into the host cells. The DNA molecule comes in the contact of the cell surface and then it is taken up by the host cells.
Transfection : Transfection is the transfer of foreign DNA into cultural host cells mediated through chemicals. This method is used for the transfer of foreign DNA in the host cell. The recipient host cells are overlayed by this mixture. Consequently, the foreign DNA is taken up by the host cell.
(c) Single cell protein : The SCP is basically a non-pathogenic, fast growing microbial biomass rich in high quality of protein and can be commercially produced throughout the year and independent of climate (except algal process).
Example : Mushrooms and yeasts are good source of vitamin B complex.
Question 3.
(a) Name any four vectors used in gene cloning technique. Also, state the unique properties of each of them. [4]
(b) With reference to tissue culture, explain the importance of: [4]
(i) Triploid plants
(ii) Haploid plants
(c) Give two uses of site directed mutagenesis. [2]
Answer:
(a) Vectors are tools used by molecular biologists to insert genes or pieces of foreign DNA into host cells. The four main types of vector commonly used include plasmids, bacteriophages, cosmids and yeast artificial chromosomes (YAC’s).
Plasmid: It is an extra chromosomal circular DNA molecule that independently replicates inside the bacterial cell and some yeasts; cloning limit: 100 to 10,000 base pairs or 0.1-10 kilobases (kb).
Phage : Designed bacteriophage lambda (λ) and M13 ; linear DNA molecules, whose region can be replaced with foreign DNA without disrupting its life cycle; cloning limit: 8-20 kb. M13 is a filamentous phage which infects E-coli. cloning limit: 10 kb.
Cosmids: A constructed extrachromosomal circular DNA molecule that combines features of plasmids and (cos) sites of phage; cloning limit: 45 kb.
Yeast Artificial Chromosomes (YAC): An artificial chromosome that contains telomeres, origin of replication, a yeast centromere, restriction enzymes sites and a selectable marker for identification in yeast cells ; cloning limit: 1Mb.
(b) (i) Applications / Importances of triploid plants :
- Autotriploidy induces gigas effect i.e., large-sized leaves, flowers, fruits and seeds.
- Autotriploids are sterile (with faulty pairing and distribution of chromosomes and abnormal gametes) and reproduce vegetatively only. They are useful for the production of seedless plants.
(ii) Applications / Importances of haploid plants :
- Haploid plants have been used in breeding programmes of rice, wheat, barley, Brassicaspps, tobacco, potato, etc.
- A doubled haploid strain are produced in only two years a net saving of 4 years for the production of homozygous lines by selfing or close breeding of both self and cross pollinated crops. They may be released directly as a variety if it performs well in yield trials.
- Haploid plants are most suitable for the study of recessive mutations.
(c) Uses of Site-directed mutagenesis
- To study the protein structure, gene expression, gene regulation and function relationships.
- To make specific and intentional changes to the DNA sequence of a gene and any gene products.
Question 4.
(a) Discuss the following innovations in biotechnology : [4]
(i) Recombinant hepatitis B vaccine
(ii) Tomato fruit with delayed ripening
(b) Explain the method used for DNA sequencing by chemical degradation technique. [4]
(c) Mention the principle involved in the freeze preservation of germplasm. [2]
Answer:
(a) (i) In July of 1986, the food and drug administration (FDA) approved the first genetically engineered vaccine for humans : Hepatitis B vaccine. Hepatitis B vaccine is developed for the prevention of hepatitis B virus infection. A microorganism, e.g., yeast for expression of hepatitis B surface antigen (HBsAg) is used as vaccine against hepatitis B virus.
The vaccine contains one of the viral envelop proteins (HBsAg). The gene coding this protein is identified and integrated into a suitable expression vector and introduced into a suitable host where the protein is produced in large quantity. Protein is then isolated and purified from host cell and used in preparation of vaccine.
(ii) A plant with delayed fruit ripening
‘Flavr Savr’ variety of tomato:
In transgenic tomato, the expression of gene for the production of polygalacturonase was blocked as this enzyme degrade pectin. In the absence of enzyme, pectin degradation is stopped and the fruit remains fresh for long time. It retains flavour, has superior taste and higher quantity’ of total soluble solids.
(b) The structure of DNA (e.g., gene insert, a recombinant plasmid or entire genome) can be analysed by determining the nucleotide sequences. In molecular cloning, the information of nucleotide sequences are essential. In 1965, Robert Holley and his research group at Cornell University completely sequenced nucleotides of tRNAala (tRNA for yeast alanine). In 1977, the following two methods were developed.
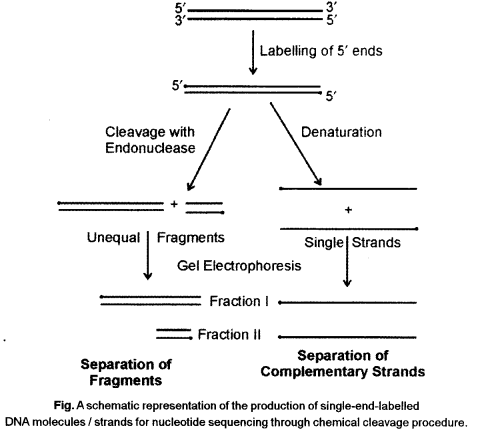
Allan Maxam and Walter Gilbert developed a chemical method of DNA sequencing. In this method, end-labelled DNA is subjected to base specific cleavage reaction before gel separation. In routine sequencing of DNA this method is not commonly followed. In the same year (1977) Frederick Sanger and co-workers developed an enzymatic method of DNA sequencing. It is also called dideoxynucleotide chain termination method because dideoxynucleotides are used as chain terminator to produce a ladder of molecules.
Maxam and Gilbert’s Chemical Degradation Method
In this method, DNA sequencing involves the following steps :
- Labelling of 3 ’ends of DNA with isotopic phosphorus (32P).
- Separation of two strands labelled at 3 ’ ends.
- Separation of mixture in four sets, each treated with a different reagent which can degrade only G or C, or A and G or T and C.
- Electrophoretic separation of each sample in four different gel.
- Autoradiography of gels and determination of the sequence from position of bands in four lanes of gel.
(c) Freezing: When cells or whole tissues are preserved by cooling to low sub-zero temperatures, such as (typically) 77 K or -196°C (the boiling point of liquid nitrogen). At these low temperatures, any biological activity, including the biochemical reactions that would lead to cell death, is effectively stopped.
Storage : The frozen cells and tissues are stored in a liquid nitrogen refrigerator, the temperature must not be rise above -130°C otherwise ice crystals may be formed.
Thawing: Thawing of the frozen materials is achieved by plunging the vials into water at 37-40°C (thawing rate of 500°-700°C/min) for 90 seconds. The material is then transferred into an ice bath till it is recultured.
Reculture : Materials subjected to cryopreservation may show some special requirements during reculture. For example, shoot-tips preserved seedlings of tomato required GA3 for developing into shoots. Similarly survival of carrot plantlets was generally improved by activated charcoal.
Question 5.
(a) Explain the principle and any two uses of each of the following biochemical techniques : [4]
(i) Partition Chromatography .
(ii) X-Ray Crystallography.
(b) Enumerate the steps involved in Southern blotting technique. Also, write two applications of this technique. [4]
(c) Mention any two methods used in the isolation of single cells from plant organs. [2]
Answer:
(a) (i) Partition Chromatography: Partition chromatography is the process of separation whereby the components of the mixture get distributed into two liquid phases due to differences in partition coefficients during the flow of mobile phase in the chromatography column.
Here the molecules get preferential separation in between two phases, i.e., both stationary phase and mobile phase are liquid in nature. So molecules get dispersed into either phases preferentially. Polar molecules get partitioned into polar phase and vice-versa. This mode of partition chromatography applies to liquid-liquid, liquid-gas chromatography and not to solid-gas chromatography. Because partition is the phenomenon in between a liquid and liquid or liquid and gas or gas and gas. But not in solid involvement.
Uses:
- It is used to separate the amino acids.
- It is extensively used for the study of lipid-soluble substances.
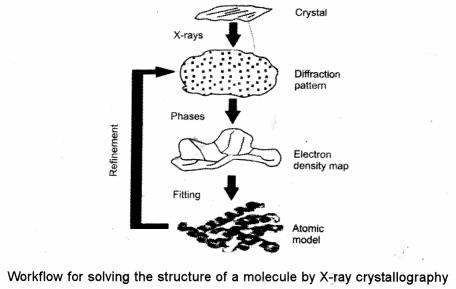
(ii) The technique of single-crystal X-ray crystallography has three basic steps :
The first and often most difficult step is to obtain an adequate crystal of the material under study. The crystal should be sufficiently large (typically larger than 100 microns in all dimensions), pure in composition and regular in structure.
In the second step, the crystal is placed in an intense beam of X-rays, usually of a . single wavelength (monochromatic X-rays), producing the regular pattern of reflections.
As the crystal is gradually rotated, previous reflections disappear and new ones appear; the intensity of every spot is recorded at every orientation of the crystal. Multiple data sets may have to be collected, with each set covering slightly more than half a lull rotation of the crystal and typically containing tens of thousands of reflection intensities.
In the third step, these data combined computationally with complementary chemical information to produce and refine a model of the arrangement of atoms within the crystal. The final, refined model of the atomic arrangement now called a crystal structure is usually stored in a public database.
Uses:
- It is used to determine the atomic and molecular structure of a crystal.
- It is also used in X-ray diffraction to rotate the samples.
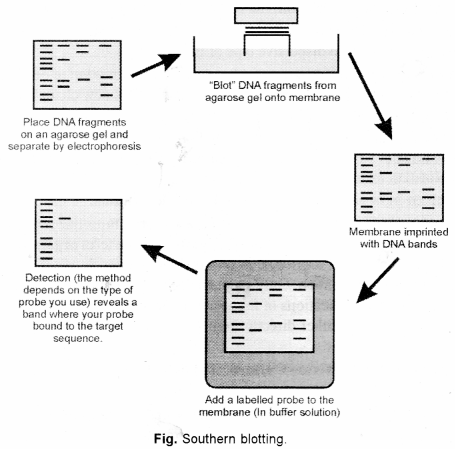
(b) Southern blotting is a technique for transfer of DNA molecules from an electrophoresis gel to a nitrocellulose or nylon membrane, and is carried out prior to detection of specific molecules by hybridization probing.
In this technique, DNA is usually converted into conveniently sized fragments by restriction digestion and separated by gel electrophoresis, usually on an agarose gel. The DNA is denatured into single strands by incubation by alkali treatment.
The DNA is transferred to a nitrocellulose filter membrane which is a sheet of special blotting paper. The DNA fragments retain the same pattern of separation they had on the gel. This process is called blotting.
The nitrocellulose membrane is now: removed from the blotting stack.
The blot is incubated with many copies of a radioactive probe which is single-stranded DNA. This probe detect and identify base pairs with its complementary DNA sequence and bind to form a double-stranded DNA molecule. The probe cannot be seen but it is either radioactive or has an enzyme bound to it (e.g., alkaline phosphatase or horseradish peroxidase). This step is known as hybridization reaction.
The location of the probe is revealed by incubating it with a colorless substrate that the attached enzyme converts to a coloured product that can be seen or gives off light which will expose X-ray film. If the probe was labelled with radioactivity, it can expose X-ray film directly. The images of radioactive probe are revealed as distinct bands on the developed X-ray film.
Applications of Southern blotting
- It is used in DNA fingerprinting.
- Identify mutation, deletion and gene rearrangement.
- Prognosis of cancer and prenatal diagnosis of genetic disease.
(c) Methods of single cell isolation from plant organ: Leaf mesophyll tissue and callus are the most suitable materials to isolate a single cell. Leaf mesophyll tissue contains a homogeneous population of cells. There are two methods described for isolation of a single cell:
Mechanical Method: Mechanical isolation involves tearing of surface sterilized explants to expose the cells followed by scrapping of the cells with a fine scalpel to liberate the single cells without any damage. These cells are dipped in liquid medium.
About 10 grams of leaves are macerated in 40 ml of buffered medium using pestle and mortar. The homogenate is filtered through muslin cloth. Cells are w ashed by centrifugation at low speed. Cells are collected and debris are removed.
Enzymatic Method : Using this method, maximum amount of cells can be isolated with minimum damage and injury in the cells. It is accomplished by providing osmotic protection to the cells. The enzyme (pectinase/macerozyme) degrades the middle lamella and cell walls of the parenchy matous tissue as a result, individual cells are set free. Osmotic protection is provided to the cells while the enzyme macerozyme degrades the middle lamella and cell w all of the paranchymatous tissues.
Question 6.
(a) State the role of any four enzymes involved in the process of DNA replication. [4]
(b ) Outline the basic steps involved in the in vitro regeneration of a complete plant from a single cell. [4]
(c) Name any two biochemical techniques based on solubility. [2]
Answer:
(a) Enzymes taking part in Replication
Four different enzymes have been reported which are given below :
Exonuclease : This enzyme breaks down the hydrogen bonds coupling nitrogenous bases in DNA molecules so that both the chains get apart during replication.
Endonuclease: This enzyme breaks down the chain or chains of DNA molecule. Whenever it appears, it results in dispersive type of replication.
Polymerase or replicase enzyme: It catalyses the formation of one polynucleotide chain that is a copy of another (complementary chain). This new chain is synthesized from the raw material already present in nucleoplasm.
Ligase: This enzyme is used in the formation of bonds between the nitrogenous bases such as A = T and C = G. Whenever the appearance of the enzyme is delayed, conservative type of replication results.
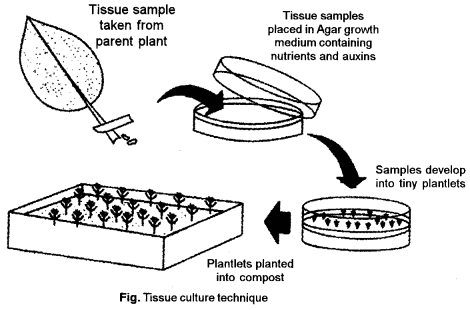
(b) Steps involved in in vitro regeneration of a complete plants.
Regeneration refers to the development of organised structures like roots, shoots, flower buds, somatic embryos (SE’s), etc. from cultured cells/tissues; the term organogenesis is also used to describe these events. Root regeneration occurs quite frequently, but it is useful only in case of shoots and embryo germination. Only shoot and SE regeneration’s give rise to complete plants, which is essential for applications of tissue culture technology in agriculture and horticulture. Regeneration may occur either directly from the explant or may follow an intervening callus phase.
Basic technique of Plant Tissue Culture: The basic technique of plant tissue culture involves the following steps :
Preparation and Sterilisation of Suitable Nutrient Medium: Suitable nutrient medium as per objective of culture is prepared and transferred into suitable containers. Culture- medium is rich in sucrose, minerals, vitamins, and hormones. Yeast extract, coconut milk, are also added. The culture is completely sterilized in an autoclave.
Selection of explants : Selection of explants such as shoot tip should be done.
Sterilisation of explants: Surface sterilisation of the explants by disinfectants (eg., sodium hypochlorite or mercuric chloride) and then washing the explants with sterile distilled water is essential.
Inoculation: Inoculation (transfer) of the explants into the suitable nutrient medium (which is sterilised by autoclaving to avoid microbial contamination) in culture vessels under sterile conditions is done.
Incubations : Growing the culture in the growth chamber or plant tissue culture room, having the appropriate physical condition (i.e., artificial light; 16 hours of photoperiod), temperature (- 26°C) and relative humidity (50-60%) is required.
Regeneration : An unorganized mass of cells developing from explants is called callus. The callus gives rise to embryoids which can develop into whole plant if the medium is provided with proper concentration of hormones. This property of developing every somatic cell into a full fledged plant is called totipotency. Regeneration of plants from cultured plant tissues is carried out.
Hardening: Hardening is gradual exposure of plantlets to an environmental conditions.
Plantlet transfer : After hardening, plantlets are transferred to the greenhouse or field conditions following acclimatization (hardening) of regenerated plants.
(c) Biochemical techniques based on solubility :
- Salt precipitation
- Precipitation with organic solvent.
Question 7.
(a) Explain the operon concept of gene regulation with the help of an example. [4]
(b) Enumerate the secondary and tertiary structures of proteins. Also, mention any two important functions of proteins. [4]
(c) Mention any two potential uses of cloned stem cells. [2]
Answer:
(a) Operon is a regulatory gene system, often encoding a coordinated group of enzymes involved in a usually by interaction with and inactivation of a repressor, of trancription of the genes encoding the enzymes. Lac-operon is type of inducible pathway as the operon is switched on in response to the presence of chemical (lactose, a subtrate) called Inducer, which after coming in contact with repressor, form a non-DNA binding complex so as to free the operator gene for transcription. The enzymes formed in response to the presence of its subtrate are called Inducible enzymes.
Lac-operon of E.coli has following components.
- There are three structural genes Z, Y, A, which transcribe polycistronic /mRNA.
- Promoter gene is the site where RNA polymerase binds for transcription.
- Operator gene function as a switch for the operon. In absence of lactose, repressor binds to operator gene and the RNA polymerase cannot move from promoter gene and thus there is no transcription.
- i-gene is a regulator gene and synthesises a repressor protein. In the presence of lactose (inducer), the repressor bind to it and become inactive repressor, which does not bind to operator gene and RNA polymerase from promoter gene move to structural genes to help transcription.
Example: The lactose operon is an example of negative regulation. When E. coli cells are growing in a medium containing lactose as the only carbon source, some of the lactose is converted to allolactose. Allolactose acts as an inducer and turns on the lac-operon. In the presence of glucose and the absence of lactose, allolactose is not produced and the lac operon is turned off. Since glucose is a readily available energy source, cells won’t use lactose until they consume all the glucose. When both glucose and lactose are present in the medium, the operon is also turned off. In this case, the operon is regulated by catabolite repression.
(b) Secondary Structure (2° structure) : It is development of new stearic relationships amongst the amino acids for protecting their peptide bonds through formation of intrapolypeptide and interpolypeptide hydrogen bonds. Secondary structure is of three types—a-helix, p-pleated and collagen helix. The prefixes a and p signify the first and second types of secondary structure discovered by Pauling and Corey (1951).
α-Helix: The polypeptide chain is spirally coiled, generally in a clockwise or right-handed fashion (Fig.). There are 3.6 amino acid residues per turn of the spiral. The spiral is stabilized by straight hydrogen bonds between imide group (-NH-) of one amino acid and carbonyl group (—CO—) of fourth amino acid residue. In this way, all the imide and carbonyl groups become hydrogen bonded. R-groups occur towards the outer side of a-helix. a-helix is the final structure in certain fibrous proteins, e.g., keratin (hair, nail, horn), epidermis (skin).
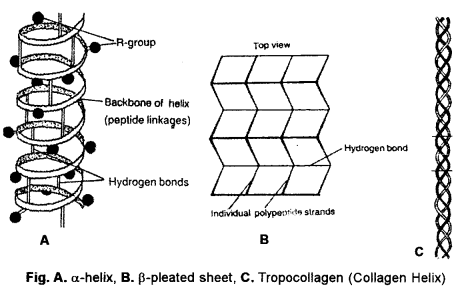
β-Pleated Sheets : Two or more polypeptide chains come together and form a sheet. Condensation is little. However, twisting does occur. The same polypeptide may fold over itself to form two strands for β-pleating. Adjacent polypeptide chains may occur in parallel (e.g., p-keratin) or anti-parallel (e.g., silk fibrin). Straight hydrogen bonds occur between imide (-NH-) group of one polypeptide and carbonyl (-CO-) group of adjacent polypeptide. Cross-linkages help in stabilization of β-pleated sheets.
Collagen Helix : Collagen has a large amount of glycine (25%) and proline (and hydroxyproline, 25%). It cannot form a-helix due to them. Three of its polypeptide each having about 1000 amino acid residues, come together with each forming an extended left-handed helix. They run parallel, form a right-handed super-helix that is stabilised by hydrogen bonds amongst the three. The triple helix of collagen is often called tropo- collagen. Its one end is stabilised by -S-S- linkages amongst the three chains. Collagen occurs in those tissues where extensibility is limited, e.g., connective tissue, tendons, bones.
Tertiary Structure (3° Structure) :
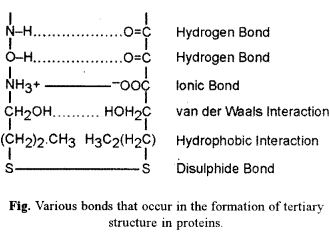
Tertiary (3°) structure is bending and folding of secondary strand (2°) of polypeptide (a-helix or β-pleated sheet) in such a way as to form a compact structure with functional sites being established over its surface by coming together of polar regions of specific amino acids. Hydrophobic parts of amino acids generally pass into the interior of protein. When both a-helix and β-pleating occur in the same polypeptide, tertiary structure segregates them into specific areas. Tertiary structure is stabilised by five types of bonds :
- Disulphide Bonds : Between hydrogen sulphide groups of two cysteine or methionine amino acids.
- Hydrogen Bonds : They occur between hydrogen and oxygen atoms of various groups like -NH OC- or -OH OC-.
- Ionic Bonds : They appear between oppositely charged ionised groups of two amino acids, e.g., NH3+ and -COO .
- Hydrophobic Interactions: They are not true bonds but involve coming together of non-polar R-groups of two amino acids. Hydrophobic interactions are important in compaction of protein as they exclude water molecules in the area of their occurrence.
- van derWaals Interactions : They develop between two closely placed polar groups through charge fluctuations between the two.
Functions of Proteins:
- Some proteins as hormones regulate many body functions. For example, the hormone insulin is a protein. It regulates sugar level in the blood.
- Some proteins as enzymes catalyse or help in biochemical reactions. For example, pepsin and trypsin.
(c) (i) Stem cells can be modified to bone, muscle, cartilage and other specialized types of cells, they have the potential to treat many diseases, including Parkinson’s, Alzheimer’s, diabetes and cancer.
(ii) New medications could be tested for safety on specialized cells generated in large numbers from stem cell lines.
Question 8.
(a) What is bioinformatics ? Mention its significant applications.
(b) Name the first bio technologically created mammalian clone and also explain the methodology involved in it.
(c) Define the term semi-gamy.
Answer:
(a) Bioinformatics is the application of computer technology to the management of biological information. Computers are used to gather, store, analyze and integrate biological and genetic information which can then be applied to gene-based drug discovery and development. Bioinformatics uses many areas of computer science, statistics, mathematics and engineering to process biological data.
Applications of Bioinformatics:
- It is used to search the genome of thousands of organisms, containing billions of nucleotides. These programs would compensate for mutations (exchanged, deleted or inserted bases) in the DNA sequence, in order to identify sequences that are related, but not identical.
- It is used for measuring biodiversity.
- The expression of many genes can be determined by this process.
- Bioinformatic techniques have been applied to explore various steps in the analysis of regulation.
- In order to trace the homology of newly identified protein.
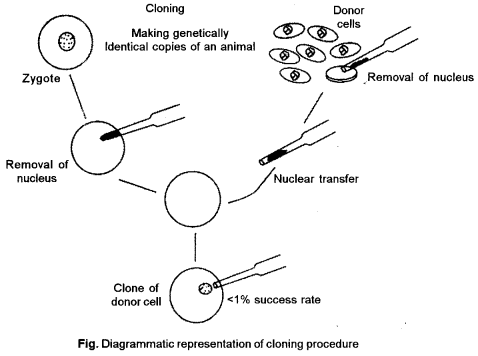
(b) Dolly (sheep) is the first cloned sheep. The methods involved in creation of dolly are :
To Isolate donor nucleus: Firstly isolate the nucleus from a somatic (non-reproductive) cell of a adult donor sheep. The nucleus contains the complete genetic material of the organism. A micropipette is used to capture the nucleus and remove it from the cell.
Recovery of unfertilized eggs: Remove some unfertilized egg cells (reproductive) from a female sheep. Many eggs are needed since not all of them will survive the various steps of cloning.
Remove the egg’s nucleus: Nucleus is removed from egg which contains only one-half of the sheep’s genetic material.
Insert donor nucleus : Insert the nucleus, with its complete genetic material, isolated from the donor mammal in Step 1 into the egg cell that has no nuclear material. The egg’s genetic material now contains all traits from the donor adult. This egg is genetically identical to the donor adult.
Place the egg into womb : Transplant the eggs into a female sheep’s womb. Those eggs that survive will continue to develop into embryos. The egg matures in the womb. The offspring dolly is born, a clone (genetically identical) of the donor sheep.
After cloning was successfully demonstrated through the production of Dolly, many other large mammals have been cloned, including horses and bulls. The attempt to clone argali (mountain sheep) did not produce viable embryos. The reprogramming process cells need to go through during cloning is not perfect and embryos produced by nuclear transfer often show abnormal development.
(c) Semigamy: It is a type of facultative apomix in which the male sperm nucleus does not fuse with the egg nucleus after penetrating the egg in the embryo sac. Subsequent development can give rise to an embryo containing haploid chimeral tissues of paternal and maternal origins. In cotton, the semigametic phenomena was first reported by Turcotte and Feaster (1963), who developed the Pimaline 57-4 that produced haploid seeds at a high frequency.
Question 9.
(a) Write short notes on : [4]
(i) Protein databases
(ii) Hardware and Software of computers
(b) Name any four resources obtained from NCBI and give their uses. [4]
(c) Mention any two types of sequence analysis used in bioinformatics. [2]
Answer:
(a) (i) Protein Databases: The protein database is a collection of sequences from several sources, including translations from annotated coding regions in GenBank, RefSeq and TPA, as well as records from SwissProt, PIR, PRF, and PDB. Protein sequences are the fundamental determinants of biological structure and function. Protein databases are more specialized than primary sequence databases. They contain information derived from the primary sequence databases.
Example : PIR, TIGR, PFam, PRINTS, ProDom, ProSite, ALIGN, SWISS-PROT & TREMBL, etc.
(ii) Hardware and Software of computers
Hardware: Hardware is the collection of physical components that constitutes a computer system including peripherals such as the monitor, mouse, keyboard, computer data storage, hard drive disk (HDD), system unit (graphic cards, sound cards, memory motherboard and chips), etc., all of which are physical objects that can be touched. In contrast, software is instructions that can be stored and ran by hardware.
Software: Written programs or procedures or rules and associated documentation pertaining to the operation of a computer system and that are stored in read/write memory. Bases on their goals, they could be Application software, System software, Malicious software or malware, etc.
(b) The NCBI located at National library of Medicine was established in Bethesda, Maryland in 1988 through legislation sponsored by Senator Claude Pepper.
Useful resources:
- Genome sequencing data in GeneBank.
- An index of biomedical research articles in PubMed Central and PubMed.
- Entrez search engine, used to access literature (abstracts), sequence and structure databases.
(c) PDB (Protein Data Bank): This database has sequence of those protein whose 3 -D structures are known.
Source : NCBI-U.S.A.; EBI, UK.
Blast (Basic Local Alignment Search Tool): Blast is a family of user-friendly sequence similarity search tools on the web. The Blast server is supported through NCBI (National Centre for Biotechnology Information) U.S.A. This tool is designed to identify potential homologues for a given sequence. It can analyse both DNA and protein sequences. A local alignment finds the optimal alignment between subregions or local regions of specified sequences.
