ISC Biotechnology Previous Year Question Paper 2013 Solved for Class 12
Maximum Marks: 80
Time allowed: Three hours
- Candidates are allowed additional 15 minutes for only reading the paper. They must NOT start writing during this time.
- Answer Question 1 (Compulsory) from Part I and five questions from Part II, choosing two questions from Section A, two questions from Section B and one question from either Section A or Section B.
- The intended marks for questions or parts of questions are given in brackets [ ].
- Transactions should be recorded in the answer book.
- All calculations should be shown clearly.
- All working, including rough work, should be done on the same page as, and adjacent to the rest of the answer.
Part-1
(Answer all questions)
Question 1.
(a) Mention any one significant difference between each of the following : [5]
(i) Gene and Genome
(ii) Multi potent cell and Uni potent cell
(iii) Galactose and Glycine
(iv) Batch culture and Continuous culture
(v) Coding region and Non-coding region
(b) Answer the following questions : [5]
(i) Name the enzyme used in PCR. What is the source of this enzyme?
(ii) Why is Bt-cotton resistant to boll worm?
(iii) Mention any two methods of ex-situ conservation of germplasm.
(iv) What is Proteomics?
(v) Glucose and fructose have the same chemical formula (C6H1206), yet they differ in chemical properties. Why ?
(c) Write the Ml form of the following : [5]
(i) GDB
(ii) PIR
(iii) YAC
(iv) NCBI
(v) ddNTP
(d) Explain briefly : [5]
(i) Bacterial Artificial Chromosome
(ii) Vascular differentiation
(iii) Phenylketonuria
(iv) Quartemary protein
(v) Designer oils
Answer:
(a) (i) Gene: Gene is the unit of the genome, consisting of a sequence of DNA that occupies a specific position (locus) on a chromosome and determines a particular characteristic in an organism.
Genome: Genome is the total genetic information or all the genes contained in a haploid set of chromosomes in eukary otes, in a single chromosome in bacteria, or in the DNA or RNA of viruses.
(ii) Multipotent: These cells have the ability to differen-tiate into many of the various type of specialized cell types and can develop into any cell of a particular group or type. e.g., umbilical cord stem cells.
Unipotent: These cells can undergo unlimited reproductive divisions, but can only differentiate into a single type of cell or tissue, e.g., skin cells.
(iii) Galactose: It is a part of disaccharide that is made- up of two sugars. It is found in milk alongwith glucose. Galactose does not occur freely in nature. It is produced in the body during the digestion of disaccharide lactose.
Glycine: Glycine is a neutral amino acid and one of the 20 building blocks of protein. It is a non-essential amino acid, used in purine synthesis, and is a neurotrans¬mitter.
(iv) Batch culture: It is a type of culture in which nutrients are fed continously depending upon the amount consumed without removing growth products.
Continuous culture: It is a open type of culture in which nutrients are supplied from time to time alongwith removal of product in same volume.
(v) Coding region: Coding region (exon) is a part of the DNA that actually codes for a protein.
Non-coding region: Non-coding region (introns) is that part of DNA that does not code directly for a protein.
(b) (i) The enzyme used in PCR is TAQ – DNA polymerase I and the source of this enzyme is Thermus aquaticus.
(ii) Bt-co.tton is an insect-resistant Genetically Modified (GM) variety of cotton seed, which contains a cry gene from Bacillus thuringiensis to kill the bollworm.
(iii) Ex-situ conservation of germplasm refers to maintaining or conserving the germplasm of organism outside their natural seed habitats. Two methods of ex-situ germplasm conservation are seed banks, botanical gardens, zoological parks etc.
(iv) Proteomics is the study of entire complement of proteins, particularly their structures and functions on the large scale. The term “proteomics” was first coined in 1997 and used to make an analog}’ with genomics, the study of the genes. The word “proteome” is derived from “protein’ and “genome”, and this was coined by Marc Wilkins in 1994. It is constantly changing due to intracellular and extracellular factors.
(v) Both Glucose and fructose have the same chemical formula, but they are different because of the different arrangement of the atoms within the molecules. Glucose is an aldose with a -CHO group at position 1 while fructose is a ketose with a -C = O at position 2.
(c) (i) Genome Data Base
(ii) Protein Information Resource
(iii) Yeast Artificial Chromosome.
(iv) National Centre for Biotechnolog} Information
(v) Dideoxynucleoside triphosphate.
(d) (i) Bacterial artificial chromosome (B AC) is a cloning l ector construct, based on a fertility plasmid (or F-plasmid). which is used for transforming and cloning in bacteria, usually E.coli. An ori gene for maintainance of F factor, a selectable marker and many restriction sites for insertion of foreign DNA. The bacterial artificial chromosome’s usual insert size is 300 to 350 kbp.
(ii) Vascular tissues are complex tissues, each consisting of a number of different types of cells. Vascular differentiation refers to the process by which different types cell types arise from precursor cells and become different in structure and function from each other.
(iii) Phenylketonuria is the recessive genetic disorder caused by the absence of the enzyme phenylalanine hydroxylase which catalyzes the conversion of phenylpyruvic acid into hydroxyphcnyl pyruvic acid. It is caused due to mutation of gene.
(iv) Quartemary proteins are the multimeric proteins i.e.. proteins hav ing more than two or more polypeptide chains which are linked to form quartemary structure, e.g.. Haemoglobin.
(v) Designer Oil: “Designer oil” that reduces LDL (‘bad“) blood cholesterol levels in humans and increases energy expenditure which may prevent people from gaining weight. The oil incorporates a phytosterol-based functional food ingredient Phytrol (TM) from Forbes into oil using proprietary technology.
Part-II
(Answer any five questions)
Question 2.
(a) Give a comparative account of DNA and RNA on the basis of their following characteristics: [4]
(i) Chemical composition and structure
(ii) Location and function
(b) Mention the uses of the following in genetic engineering techniques : [4]
(i) Shuttle vectors and Expression vectors
(ii) Restriction endonucleases
(c) What is electroporation ? [2]
Answer:
(a) (i) DNA:
- DNA has 2-Deoxyribose sugar.
- It contains cytosine and thymine as pyrimidine.
- It has a double stranded helix struc-ture.
RNA:
- RNA has ribose sugar.
- It contains cytosine and uracil as pyrimidine.
- It has a single stranded helix.
(ii) DNA:
- DNA occurs in the nucleus chloroplast and mitochondria of cell.
- It controls transmission of hereditary characters.
RNA:
- RNA occurs in cytoplasm of the cell.
- It controls the synthesis of proteins.
(b) (i) Shuttle vectors exist and work and allow DNA to be transferred between both prokaryotes and eukaryotes. The shuttle vector has two origins of replication i.e.. onE and oriEuk allowing replication to occur in either system/host. It “shuttles” between two different species. It can be used to perform reverse genetics, e.g.. Yeast episomal plasmid (YEP). Expression vectors allow expressing certain genes directly from their recombinant DNAs A typical expression vector will have a promoter upstream of the DNA containing the sequence to be expressed.
(ii) Restriction endonucleases are enzymes that cleave DNA at specific nucleotide sequences. The sequence recognized is often four to six nucleotides long. For example, the restriction endonucleases Eco RI recognize the sequence. GAATTC.
(c) Electroporation is a mechanical method used to introduce polar molecules into a host cell through the cell membrane. In this procedure, a brief exposure to a high electric voltage pulse temporarily disturbs the phospholipid bilayer, allowing introduction of molecules like DNA to pass into the cell.
Question 3.
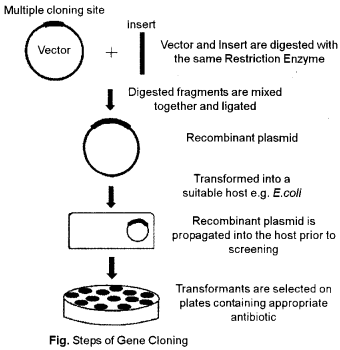
(a) What is gene cloning ? Mention the steps involved in this process
(b) Explain the following :
(i) Acidic and basic amino acids
(ii) Phospholipids and glycolipids
(c) State any four objectives of germplasm conservation.
Answer:
(a) The process of formation of similar copies of a desired gene is called gene cloning. Gene cloning is the technique of recombinant DNA technology’ in which a desired gene of interest having a characteristic feature is cloned. Gene cloning involves the replication of DNA fragments by the use of self-replicating vector’s genetic material for its multiplication, expression or integration into host chromosome.
Steps involved in gene cloning :
- In cloning a gene the first step is to isolate the DNA segment from the organism that contains the gene of interest.
- Remove the gene of interest from the DNA, by using restriction enzymes or by PCR.
- Vector is also treated with same restriction enzyme, to cleave it. Vector come to possessing single strand at the ends called stick}’ ends.
- Then the enzy me DNA ligase is used to insert the gene of interest to be cloned into the plasmid. Vector having sicky ends to form recombinant DNA.
- The plasmid or vector acts as a vehicle that transports the desired gene into a host cell, the process is known as transformation.
- Now, these recombinant plasmids are inserted into bacterial host cells, where they replicate to amplify the desired gene, the process is called gene cloning.
- Now the cell can be plated out on an agar medium. The colony of cells containing the desired cloned gene can be identified and isolated.
(b) (i) Amino acids are the basic structural unit of all proteins. A free’ neutral amino acid (a single amino acid) always has an amino group -NH2. a carboxyl group -COOH, hydrogen -H and a chemical group or side chain -”R”.
Acidic amino acid :
Two amino acids have acidic side chains at neutral pH. These are aspartic acid or aspartate (Asp) and glutamic acid or glutamate (Glu). Their side chains have carboxylic acid groups whose pKa’s are low enough to lose protons, becoming negatively charged in the process. Such amino acids are highly polar.
Basic amino acid :
Three amino acids have three basic side chains at neutral pH. These are arginine (Arg), lysine (Lys). and histidine (His). Their side chains contain nitrogen and resemble ammonia, which is a base. Their pKa’s are high enough that they tend to bind protons, gaining a positive charge in the process.
(ii) Phospholipids are the phosphorylated triglyceride lipids in which one fatty acid is replaced by phosphate group added by phosphorylation. Glvcolipids are the glycosylated lipids in which sugar residue galactose or carbohydrate molecule is added by glycosylation. Phospholipids and glycolipids both are the derivatives of lipids. They form an essential component of cell membrane which plays a role in structure, maintenance and also help in eliciting certain immune reactions.
(c) Objectives of Germplasm Conservation :
- Conservation of rare germplasm arising through somatic hybridization.
- Storage of pollen for enhancing longevity.
- Maintainance of recalcitrant seeds.
- To develop genes for adaptations / endurance to varying, unfavorable biotic/abiotic stresses / environments.
- To develop high yielding varieties.
Question 4.
(a) Why are enzymes temperature sensitive ? Briefly explain the mode of action of enzymes on their substrate. [4]
(b) How is the hormone insulin synthesized, using genetic engineering technique ? State two ways in which this technique is better than the techniques used earlier. [4]
(c) What is a supra-molecular assembly ? [2]
Answer:
(a) Enzymes are temperature sensitive because almost all enzymes are proteins have tertiary structure and only function in a specific range of temperature. Exposing enzymes to high temperature break bonds and can cause them to denature, which alter the shape of the enzyme. Due to change in shape the substrate no longer ‘fits’ inactive site of the enzyme and can no longer function as normal.
Mode of enzyme action : It can be explained by this models :
Lock and key mechanism : This model was proposed by Emil Fisher in 1898. It is also called the template model. According to this model the union of the substrate and the enzyme takes place at the active site, more or less in a manner in which a key fits in a lock and results in formation of an enzyme substrate complex. As the two molecules are involved, this hypothesis is also known as the concept of inter molecular fit. The ES complex is highly unstable and almost immediately this complex breaks to produce the end product of the reaction and regenerate the free enzyme. The ES complex results in the release of energy.
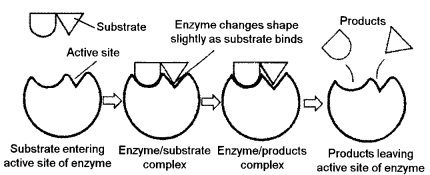
Examples:
Catalase : It catalyzes the decomposition of hydrogen peroxide into water and oxygen.
2H2O2 → 2H2O + O2
One molecule of catalyses can break 40 million molecules of hydrogen peroxide each second.
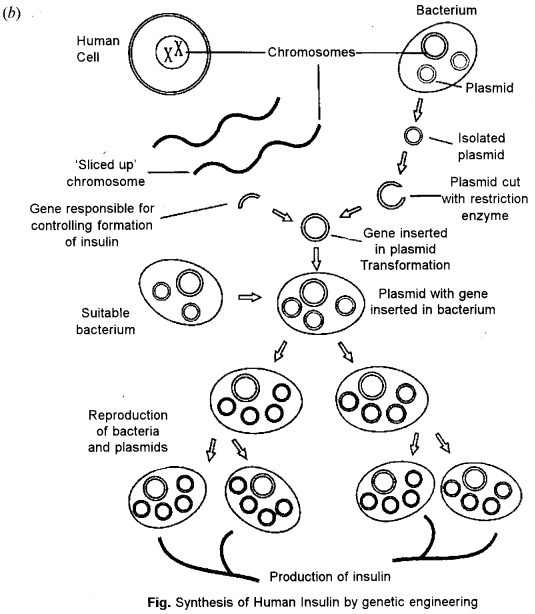
The first major medicinal product of genetic engineering is human insulin called Humulin. Insulin is a protein that acts as a hormone to stimulate uptake of blood sugar into tissues, such as the liver and the muscles.
Following are the steps which are involved in insulin synthesis :
- Isolate the gene responsible for producing human insulin protein. The gene is a part of the DNA in a human chromosome.
- Then remove a circular piece of DNA called plasmid from a bacterial cell. Special restriction enzymes are used to cut the plasmid ring open with sticky ends.
- With the plasmid ring open, the gene for insulin is inserted into the plasmid ring and the ring is closed with ligase enzyme forming recombinant DNA. This process is called recombinant technology’.
- The bacterial plasmid DNA now contains the human insulin gene and is inserted into a bacteria.
- Many plasmids with the insulin gene are inserted into many bacterial cells. When the bacterial cells reproduce by dividing, the human insulin gene is also cloned in the newly cloned cells.
- Human insulin protein molecules produced by bacteria are gathered and purified by down stream process by culturing the genetically engineered bacteria, limitless supplies of insulin may be produced.
Two ways in which genetic engineering is better than the technique used earlier:
- Insulin produced by genetic engineering is pure and has no allergic reaction.
- The human insulin is much cheaper when produced by r-DNA technology than was the insulin from cows, as it could be produced much more quickly in greater quantity.
(c) A supra molecular assembly or “super molecule” is a well defined complex of molecules held together by non-covalent bonds. Molecules are combined in the form of sphere or rod. The dimensions of supra molecular assemblies can range from nanometres to micrometers. The process by which a supra molecular assembly forms is called molecular self-assembly.
Question 5.
(a) What is plant tissue culture ? Discuss the organization of a tissue culture laboratory under the following headings: [4]
(i) Media preparation
(ii) Culture room.
(b) Explain any two methods used for the identification of recombinant host cells from the non-recombinant host cells. [4]
(c) Name any four in vivo techniques employed in haploid production. [2]
Answer:
(a) Plant tissue culture is the technique of in vitro maintenance and growth of plant cells, tissues and organs under aseptic conditions on a suitable artificial culture medium contained in small containers under controlled environmental conditions of temperature and light.
(i) Media Preparation Room : An area is required for preparation of media. In such space there should be provision for bench space for chemicals, labware, culture vessels, closures and miscellaneous equipment required for media preparation and dispensing. In this room provision is also made for placing hot plates or stirrers, pH meter, balance, waterbath, burners, oven, autoclave, culture vessel, refrigerator etc.
(ii) Culture Room : All types of cultured plant tissues are incubated under the conditions of well controlled temperature, humidity, illumination and air circulation. The culture room should have light and temperature control system. Generally temperature is maintained at 25±2°C and 20-98% relative humidity and uniform air ventilation. The cultures are grown in diffused light and darkness each for a period of 12 hours.
(b) The introduction of the recombinant DNA in to a suitable host cell is followed by the selection of those cells, which contain the recombinant vectors. There are various selection methods that are based on the expression or non-expression of some of the traits present in the vector or alongwith the cloned gene.
Antibiotic sensitivity : Recombinant plasmid has many traits such as ori recognition site and selectable marker gene. Some of these traits are resistant to certain antibiotics. If the antibiotic resistant gene is present alongwith the cloned gene, it is very easy to select the recombinant transformants directly on a medium supplemented with respective antibiotic.
In most of the cases there are two stages of selection. First is the selection on the basis of . transformed cells i.e., the cells that have taken a plasmid. The second one is to identify the transformed cells that have the recombinant plasmid. The presence of a desired DNA insert can be confirmed either by isolating the recombinant plasmids and digesting it with the same restriction enzyme used for making the recombinant vectors, by PCR, by southern hybridisation with DNA probes, by northern hybridisation with RNA probes and by direct DNA sequencing
lnsertional inactivation : Another method to differentiate between recombinant and non¬recombinant is on the basis of their ability to produce colour.
lnsertional inactivation: In this method, a recombinant DNA is within the coding sequence of an enzyme p-galactosidase. This results into the inactivation of enzyme which is referred to as insertional inactivation.
The bacterial colonies whose plasmids do not have insert, produce blue colour but those with an insert or the recombinant do not produce any colour and are identified as recombinant colonies.
(c) In vivo techniques employed in haploid production are gynogenesis, ovule and rogenesis, genome elimination by distant hybridisation or chemical treatment and semigamy.
Question 6.
(a) Write the principle and any two applications of each of the following biochemical techniques : [4]
(i) Ion – exchange chromatography
(ii) Gel – permeation
(b) What is a genetic code ? Enlist three important properties of genetic code. [4]
(c) What are DNA probes ? [2]
Answer:
(b) (i) Principle of Ion-exchange chromatography : It is defined as the reversible exchange of ions in solution with ions electrostatically bound to some sort of insoluble support medium. Separation is obtained since different molecules have different degree of interaction with the ion-exchanger due to difference in their charges, charge densities and distribution of charge on their surfaces. These interactions can be controlled by varying conditions such as ionic strength and pH.
An ion-exchanger consists of an insoluble matrix to which charged groups have been covalently bound. Ion exchange separations are carried out mainly in columns packed with an ion-exchanger. There are two types of ion-exchanger, namely cation and anion exchangers. Cation exchangers possess negatively charged groups and these will attract positively charged cations. Anion exchangers have positively charged groups that will attract negatively charged anions. After the ion exchange the molecules can be eluted from the matrix by selective desorption. The selective desorption can be achieved by changes in pH and /or ionic concentration or by affinity elution, in which case an ion that has greater affinity for the exchange than has the bound ion is introduced into the system.
Applications:
- Polystyrene and polyphenolic ion exchange resins are more often used to separate srhall molecule such as amino acids, small peptides, nucleotides, N-bases, cyclic nucleotides, organic acids.
- The cellulose ion exchangers are commonly used for proteins, including enzymes, polysaccharides and nucleic acids.
Principle of Gel-permeation chromatography: Gel permeation / filtration chromatography is a separation technique which uses molecular sieves, composed of neutral cross-linked carriers e.g., polymers like agarose, dextrans of different pore sizes. Therefore, it can separate macromolecule of different sizes from one another. Molecules smaller than pore size either the carrier and are retained. They are later eluded (in order of molecular size) and collected. Other names that have been suggested for this technique are : get filtration, molecular or size exclusion chromatography ; or molecular sieve chromatography.
Applications:
- Separation of polysaccharide, enzymes, antibodies and other proteins.
- Separation of non-polar species such as triglycerides in non-aqueous mobile phases.
- Used to analyse the molecular-weight distribution of organic soluble polymer.
The genetic code is called a triplet code, i.e sequence of three nitrogenous bases on m-RNA that specifies the recognition of a particular of a single amino acid. Thus, the information encoded in the sequence of nitrogenous bases must be read in groups of three, (UAC, GGC, UGC).
Three important properties:
- Triplet code : Three adjacent nitrogen bases constitute a codon which specifies the placement of one amino acid in a polypeptide.
- Start signal : Polypeptide synthesis is signaled by AUG or methionine codon and GUG — Valine codon. They have dual function.
- Stop signal: Polypeptide chain termination is signaled by three termination codons — UAA, UAG, and UGA. They do not specify any amino acid and are hence also called non-sense codon.
- Universal code : The genetic code is applicable universally i.e., the codon specifies the same amino acid from a virus to a tree or human being.
- Non-ambiguous codon : One codon specifies only one amino acid and not any other.
(c) DNA Probe : It is a solution of radioactive, single-stranded DNA or oligodeoxy nucleotides (a DNA segment of few to several nucleotides). The name probe signifies the fact that this DNA molecule is used to detect and identify the DNA fragment in the gel/membrane that has a sequence complementary to the probe. The probe hybridises with the complementary DNA on the membrane to the greater extent with a low non-specific binding on the membrane. This step is known as hybridisation reaction.
Question 7.
(a) How can the following plants be obtained, using genetic transformation techniques . [4]
(i) Drought and salinity tolerant plants
(ii) Somatic hybrids
(b) Explain the process involved in the transcription of DNA to mRNA. Also, mention any two post transcriptional changes that occur in the mRNA formed. [4]
(c) What are Okazaki fragments ? How are they joined ? [2]
Answer:
(i) Drought tolerance : Water is crucial for all living things. Plants use water as a solvent, a transport medium, an evaporative coolant, physical support, and as a major ingredient for photosynthesis. Without sufficient water, agriculture is impossible. Therefore, drought tolerance is an extremely important agricultural trait.
One way of engineering drought tolerance is by taking genes from plants that are naturally drought tolerant and introducing them to crops. The resurrection plant (Xerophyta viscosa), a native of dry regions of southernmost Africa, possesses a gene for a unique protein in its cell membrane. Experiments have shown that plants given this gene are less prone to stress from drought and excess salinity.
Some genes have been found that control the production of the thin, protective cuticle found on leaves. If crops can be grown with a thickened waxy cuticle, they could be better equipped for dealing with dryness.
Salt tolerance: Irrigation has enabled the transformation of arid regions into some of the world’s most productive agricultural areas. Excess salinity, however, is becoming a major problem for agriculture in dry parts of the world. In several cases, scientists have used biotechnology to develop plants with enhanced tolerance to salty conditions.
Researchers have noticed that plants with high tolerance to salt stress possess naturally high levels of a substance called glycine betaine. Further, plants with intermediate levels of salinity tolerance have intermediate levels, and plants with poor tolerance to salinity have little or none at all. Genetically modified tomatoes with enhanced glycinebetaine production have increased . tolerance to salty conditions.
Another approach to engineering salt tolerance uses a protein that takes excess sodium and diverts it into a cellular compartment where it does not harm the cell. In the lab, this strategy was used to create test plants that were able to flower and produce seeds under extreme salt levels. Commercially available crops with such a modification are still several years away.
(ii) Process, other than the sexual cycle has recently become available for higher plants, which can lead to genetic recombination. This non-conventional genetic procedure involving fusion between isolated somatic protoplasts under in vitro conditions and subsequent development of their product (heterokaryon) to a hybrid plant is known as somatic hybridisation.
Application of Somatic Hybridisation :
- Somatic cell fusion appears to be the only means through w hich two different parental genomes can be recombined among plants that cannot reproduce sexually (asexual or sterile).
- Protoplasts of sexually sterile (haploid, triploid, and aneuploid) plants can be fused to produce fertile diploids and polyploids.
- Somatic cell fusion overcomes sexual incompatibility barriers. In some cases, somatic hybrids between two incompatible plants have also found application in industry or agriculture.
- Somatic cell fusion is useful in the study of cytoplasmic genes and their activities and this information can be applied in plant-breeding experiments.
(b) The process of transcription: Transcription is the process of creating a messenger RNA strand from DNA, performed by the enzyme RNA polymerase, Transcription always occurs in a 5′ → 3′. direction, with polymerase moving 3′ → 5′ along the DNA strand.
Transcription Initiation : There are three steps in transcription :
Initiation : RNA synthesis begins after the RNA polymerase attaches to the DNA and unwinds it. RNA synthesis will always occur on the template strand.
Elongation : RNA polymerase unwinds the DNA double helix and moves downstream and elongates the RNA transcript by adding ribonucleotides in a 5′ → 3′ direction. Each ribonucleotide is added to the growing mRNA strand using the base pairing rules (A binds with T, G binds with C). For each C encountered on the DNA strand a G is inserted in the RNA, for each Q a C and for each T, an A is inserted. Since there is no T in RNA, U is inserted whenever an A is encountered. After RNA polymerase has passed, the DNA restores its double stranded structure.
Termination: When the mRNA is complete, the mRNA is released and the RNA polymerase releases from the DNA.
Two post transcriptional changes that occur in the mRNA formed are:
RNA transcripts eukaryotes are modified or processed, before leaving the nucleus to produce functional wRNA. It is processed in two ways :
(1) 5 ‘ capping : Capping of the pre-mRNA involves the addition of 7-methylguanosine (m7G) to the 5′ end.,
(2) 3′ polyadenylation: The pre-mRNA processing at the 3′ end of the RNA molecule involves cleavage of its 3′ end and then the addition of about 200 adenine residues to form a poly (A) tail. The cleavage and adenylation reactions occur if a polyadenylation signal sequence (5′ – AAUAAA-3′) is located near the 3′ end of the pre-mRNA molecule, followed by another sequence, which is usually (5′ -CCA-3’).
(c) Okazaki fragments are short, newly synthesized DNA fragments produced discontinously in pieces during DNA replication. They are formed on the lagging template strand and are complementary to the lagging template strand. Okazaki fragments are joined together by DNA ligase enzyme.
Question 8.
(a) What is meant by the term genomics ? Write the differences between structural genomics and functional genomics. [4]
(b) Name and explain any four methods of synchronization of cells. [4]
(c) What is meant by Expressed sequence tags ? [2]
Answer:
(a) The word ‘genomics’ has taken root from the term ‘genome’ which is an organism’s total genetic constitution mapping, sequencing and analyzing the genomic information to solve a medical, industrial or biological query. Genomics studies investigate structure and function of genes and do this simultaneously for all the genes in a genome. Genomics is broadly categorized into structural and functional genomics.
Structural Genomics : The structural genomics deals with DNA sequencing, sequence assembly, sequence organisation and management. Basically it is the starting stage of genome analysis i.e,. construction of genetic, physical or sequence maps of high resolution of the organism. The complete DNA sequence of an organism is its ultimate physical map. Due to rapid advancement in DNA technology and completion of several genome sequencing projects for the last few years, the concept of structural genomics has come to a state of transition. Now it also includes systematics and determination of 3D structure of proteins found in living cells. Because proteins in every group of individuals vary and so there would also be variations in genome sequences.
Functional Genomics: It is based on the information of structural genomics the next step is to reconstruct genome sequences and to find out the function that the genes do. This information also lends support to design experiment to find out the functions that specific genome does. The strategy of functional genomics has widened the scope of biological investigations. This strategy is based on systematic study of single gene / protein to all genes/proteins.
Therefore, the large scale experimental methodologies (along with statistically analysed / computed results) characterise the functional genomics. Hence, the functional genomics provide the novel information about the genome. This eases the understanding of genes and function of proteins, and protein interactions.
(b) Cell culture synchronization : Cells in suspension cultures vary greatly in size, shape, DNA, and nuclear content. Moreover, the cell cycle time varies considerably within individual cells. Therefore, cell cultures are mostly asynchronous.
It is essential to manipulate the growth conditions of an asynchronous culture in order to achieve a higher degree of synchronization. A synchronous culture is one in which the majority of cells proceed through each cell cycle phase (G1: S, G2 and M) simultaneously.
Synchronization can be achieved by following methods :
- Physical methods include selection by volume (size of cell aggregate.)
- Chemical methods include starvation (depriving suspension cultures of an essential growth compound and culture supplying).
- Chemical methods include inhibition (temporarily blocking the progression of events in the cell cycle using a biochemical inhibitor and then releasing the block).
(c) An Expressed Sequence Tag or EST is a short sub-sequence of a transcribed cDNA sequence represents a partial gene. They may be used to identify gene transcripts, and are instrumental in gene discovery and gene sequence determination, used in micro-arrays.
Question 9. .
(a) What is Human Genome Project ? Mention its objectives and significant achievements. [4]
(b) Write short notes on : [4]
(i) Locus – link
(ii) Microprocessor
(iii) EMBL
(iv) Taxonomy Browser
(c) What is site-directed mutagenesis ? [2]
Answer:
(a) The Human Genome Project (HGP) : This is an international scientific research project with a primary goal to determine the sequence of chemical base pairs which make-up DNA and to identify the approximately 25,000 genes of the human genome from both a physical and functional standpoint.
Benefits: The work on interpretation of genome data is still in its initial stages. It is anticipated that detailed knowledge of the human genome will provide new avenues for advances in medicine and biotechnology. A number of companies, such as Myriad Genetics started offering easy ways to administer genetic tests to a variety of illnesses, including breast cancer, disorders of homeostasis, cystic fibrosis, liver diseases and many others. Also, the etiologies for cancers, Alzheimer’s disease and other areas of clinical interest are considered likely to benefit from genome information and possibly may lead in the long term to significant advances in their management.
There are also many tangible benefits for biological scientists. For example, a researcher investigating a certain form of cancer may have narrowed down his/her search to a particular gene. By visiting the human genome database on the world wide web, this research can examine what other scientists have written about this gene, including (potentially) the three-dimensional structure of its product, its function(s), its evolutionaty relationships to other human genes, or to genes in mice or yeast or fruit flies, possible detrimental mutations, interactions with other genes, body tissues in which this gene is activated, diseases associated with this gene or other data types.
(b) Locus Link is a National Center for Biotechnology Information (NCBI) online resource. It is designed to link together related information on genetic loci and gene products from several sources.
Microprocessor : A microprocessor or processor is the heart of the computer and it performs all the computational tasks, calculations and data processing etc. inside the computer. Microprocessor is the brain of the computer. In the computers, the most popular type of the processor is the Intel Pentium chip and the Pentium IV is the latest chip by Intel Corporation. The microprocessors can be classified based on the following features.
Instruction Set: It is the set of the instructions that the Microprocessor can execute.
Bandwidth : The number of bits processed by the processor in a single instruction.
Clock Speed : Clock speed is measured in the MHz and it determines that how many instructions a processor can processed.
European Molecular Biology Laboratory (EMBL) : It was established to collect, organise and distribute data on nucleotide sequence and other information rebated to them. Nucleotide Sequence Database (also known as EMBL -Bank) constitutes Europe’s primary nucleotide sequence resource. Main sources for DNA and RNA sequences are direct submission from individual researches, genome sequencing projects and patent applications.
Taxonomy Browser: This search tool provides taxonomic information on various species. The Taxonomy database of NCBI has information (including scientific and common names) about all organisms for which some sequence information is available (over 79,000 species). The server provides genetic information and the taxonomic relationship of the species in question. Taxonomy has links with other servers of NCBI e.g., structure and PubMed.
(c) Site-directed mutagenesis is a molecular biology technique in which a mutation is created at a specific site in the DNA molecule.
