ISC Biotechnology Previous Year Question Paper 2012 Solved for Class 12
Maximum Marks: 80
Time allowed: Three hours
- Candidates are allowed additional 15 minutes for only reading the paper. They must NOT start writing during this time.
- Answer Question 1 (Compulsory) from Part I and five questions from Part II, choosing two questions from Section A, two questions from Section B and one question from either Section A or Section B.
- The intended marks for questions or parts of questions are given in brackets [ ].
- Transactions should be recorded in the answer book.
- All calculations should be shown clearly.
- All working, including rough work, should be done on the same page as, and adjacent to the rest of the answer.
Part-1
(Answer all questions)
Question 1.
(a) Mention any one significant difference between each of the following: [5]
(i) Hybrid and Cybrid.
(ii) DNA polymerase and Taq DNA polymerase.
(iii) Glycosidic bond and Peptide bond,
(iv) Oils and Waxes.
(v) Homopolysaccharide and Heteropolysaccharide.
(b) Answer the following questions : [5]
(i) What is a callus?
(ii) Name the method used for the sterilization of plant hormones and vitamins.
(iii) Why is DNA replication called semi-conservative replication?
(iv) What is a promoter gene?
(v) State two uses of stem cells.
(c) Write the full form of the following :
(i) HGP
(ii) STS
(iii) CSIR
(iv) LAF
(v) SCP
(d) Explain briefly:
(i) Amphipathic property of lipids.
(ii) Replication fork
(iii) Androgenesis
(iv) Transamination.
(v) Active site.
Answer:
(a) (i) Hybrid: Plants produced through the fusion of protoplasts of two different plant species/varieties is called hybrid.
Cybrid: Cybrid or cytoplasmic hybrid are cells or plant containing nucleus of one species but cytoplasm from both the parental species.
(ii) DNA polymerase: A DNA polymerase is an enzyme that catalyzes the polymerization of deoxyribonucleotides into a DNA strand in DNA replication.
Taq DNA polymerase: Taq polymerase, is a thermostable DNA polymerase, originally isolated from bacterium Thermits aquaticus. It is used in polymerase chain reaction.
(iii) Glycosidic bond: A glycosidic bond (C-O-C) is a type of covalent bond that joins a aldose or ketone group of a carbohydrate (sugar) molecule to another group (OH) which may or may not be another carbohydrate.
Peptide bond: A peptide bond (HN – C = 0) is a chemical bond formed between two amino acids when the carboxyl group of one amino acid molecule reacts with the amino group of the other amino acid molecule. thereby releasing a molecule of water.
(iv) Oils: Oils are the esters of unsaturated fatty acid with glycerol. Oils are liquid at room temperature and have low melting point.
Waxes: Waxes are esters of fatty acid other than glycerol. They contain one molecule of long chain fatty acid esterified with one molecule of long chain like cytyl, ceryl or mericvl, mono hydroxy’ alcohol.
(v) Homopolysaccharide: Homopolysaccharides are the complex carbohydrates formed by polymerisation of the monosaccharide monomers.
Heteropolysaccharide: Heteropoly saccharides are the complex carbohy drates w hich are produced by the condensation of more than one type of monosaccharide monomers or their derivatives e.g., chitin, agar.
(b) (i) Callus is a mass of meristematic, undifferentiated cells derived from plant tissue (explants).
(ii) The autoclaving denatures the vitamins and hormones, therefore, the solution of these compounds are sterilized by using Millipore filter paper with pore size of 0.2 micrometer diameter.
(iii) Semi-conservative is that mode of replication in which out of the two DNA strands, one is conserved strand and the other is newly synthesised.
(iv) A promoter gene is a segment of DNA containing the enzyme RNA polymerase that initiates the transcription of the genetic code.
(v) Uses of stem cells :
(1) Bone marrow transplants that are used to treat leukemia (blood cancer).
(2) Treatment for muscular dystrophy.
(c) (i) HGP: Human Genome Project.
(ii) STS: Sequence Tagged Sites.
(iii) CSIR : Council of Scientific and Industrial Research.
(iv) LAF: Laminar Air Flow.
(v) SCP: Single Cell Protein. .
(d) (i) Amphipathic nature : Most membrane lipids are amphipathic, having a non-polar end and a polar end. The amphiphilic nature of some lipids allows them to form structures such as vesicles, liposomes, or membranes in an aqueous environment.
(ii) The point at which the two strands of DNA are separated to allow replication of each strand is known as replication fork.
(iii) Androgenesis is the development of an embryo containing only paternal chromosomes due to failure of the egg to participate in fertilization.
(iv) Transamination is the process of transfer of the a-amino group from the amino acid to an a-keto acid.
(v) Active site is part of an enzyme where substrates bind and undergo a chemical reaction.
Part-II
(Answer any five questions)
Question 2.
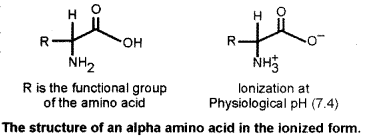
(a) Explain the general structure of an amino acid. What do you understand by essential and non-essential amino acids? [4]
(b) What are cloning vectors ? Write the main characteristics of any three types of cloning vectors. [4]
(c) What is a Codon ? Name the start codon and any one end codon. [2]
Answer:
(a) Amino acids are building blocks of macromolecular proteins. They contain amino group and carboxyl group as functional groups.
On the basis of nutritional values amino acids are of two types :
Essential amino acids : These are essential for our body but they are not synthesised inside our body are termed as essential amino acids e.g., valine, leucine, isoleucine, lysine, phenylalanine, methionine, threonine, histidine, arginine.
Non-essential amino acids : They are those which are synthesised through transformation and transamination inside our body e.g., serine, alanine etc.
(b) Cloning vector is a self-replicating DNA molecule that carries foreign DNA insert into a host cell, replicates inside a bacterial (or yeast) cell and amplify to produce many copies of itself and the foreign DNA.
Plasmid: It is an extra chromosomal circular DNA molecule that self replicates inside the bacterial cell and some yeast; cloning limit: 100 to 10,000 base pairs or 0.1-10 kilobases (kb).
Phage : Designed bacteriophage lambda (A.) and Ml 3 : linear DNA molecules, whose region can be replaced with foreign DNA without disrupting its life cycle; cloning limit: 8-20 kb. M13 is a filamentous phage which infects E-coli. Cloning limit: 10 kb.
Cosmids: A constructed extrachromosomal circular DNA molecule that combines features of plasmids and ‘cos’ site of phage ; cloning limit – 45 kb.
Yeast Artificial Chromosomes (YAC) : An artificial chromosome that contains telomeres, origin of replication, a yeast centromere, restriction enzyme site and a selectable marker for identification in yeast cells ; cloning limit: 1 Mb.
(c) Codon is a unit of genetic coding, consists of a series of three adjacent bases (triplet) in one polynucleotide chain of a DNA or RNA molecules, which codes for a particular amino acid during synthesis of proteins in a cell. For example, ATA codes for Leucine.
Start codon : The codon AUG specifies the first amino acid, methionine, in protein synthesis.
End codon : UAG also referred to as amber codon, in mRNA which terminate translation.
Question 3.
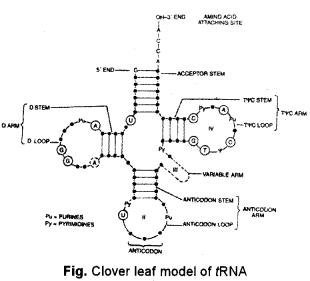
(a) Briefly explain the structure of a tRNA molecule. Mention its function during the process of protein synthesis. [4]
(b) Give the stepwise procedure of sequencing of DNA by Sanger’s method. [4]
(c) What is Totipotency ? Give an example of a Totipotent cell. [2]
Answer:
(a) Transfer RNA (tRNA) : It is also called soluble or sRNA. There are over 100 types of tRNAs. Transfer RNA constitutes about 15% of the total RNA. tRNA is the smallest RNA with 70-85 nucleotides and sedimentation coefficient of 4S. The nitrogen bases of several of its nucleotides get modified e.g., pseudouridine (φ), dihydrouridine (DHU), inosine (I). This causes coiling of the otherwise single-stranded tRNA into L-shaped form (three-dimensional, Klug, 1974) or clover-like form (two dimensional, Holley, 1965). About half of the nucleotides are based paired to produce paired stems. Five regions are unpaired of single-stranded—AA-binding site, Tig C loop, DHU loop, extra arm and anticodon loop.
- Anticodon. It is made-up of three nitrogen bases for recognizing and attaching to the codon of wRNA.
- AA-Binding site. It lies at the 3′ end opposite to the anticodon and has CCA — OH group (5′ ends bears G). Amino acid or AA-binding site and anticodon are the two recognition sites of tRNA.
- T φ C loop. It contains pseudouridine. The loop is the site for attaching to ribosomes,
- DHU loop. The loop contains dihydrouridine. It is binding site for aminoacyl synthetase enzyme,
- Extra arm. It is a variable site arm or loop which lies between T ig C loop and anticodon. The exact role of extra arm is not known.
Functions:
tRNA is adapter molecule which is meant for transferring amino acids to ribosomes for synthesis of polypeptides. There are different tRNAs for different amino acids. Some amino acids can be picked up by 2-6 tRNAs. tRNAs carry specific amino acids at particular points during polypeptide synthesis as per codons of mRNA. Codons are recognised by anticodons of tRNAs. Specific amino acids are recognised by particular activating or aminoacyl synthetase enzymes,
They hold peptidyl chains over the mRNAs. The initiator tRNA has the dual function of initiation of protein synthesis as well as bringing in of the first amino acids. There is, however, no tRNA for stop signals.
(b) DNA sequencing: It is the determination of the precise sequence of nucleotides in a sample of DNA.
Sanger dideoxy method: The most popular method for DNA sequencing is called the dideoxy method or Sanger method (named after its inventor, Frederick Sanger, who was awarded the 1980 Nobel prize in chemistry).
The Procedure : The DNA to be sequenced is prepared as a single strand.
This template DNA is supplied with
a mixture of all four normal (deoxy) nucleotides in ample quantities
- dATP
- dGTP
- dCTP
- dTTP
a mixture of all four dideoxynucleotides, each present in limiting quantities and each labeled with a tag.
- that fluoresces a different color :
- dd ATP
- dd GTP
- dd CTP
- dd. TTP
DNA polymerase 1
Because all four normal nucleotides are present, chain elongation proceeds normally until, by chance, DNA polymerase inserts a dideoxy nucleotide (shown as colored letters) instead of the normal deoxvnucleotide (shown as vertical lines). If the ratio of normal nucleotide to the dideoxy versions is high enough, some DNA strands will succeed in adding several hundred nucleotides before insertion of the dideoxy version halts the process.
At the end of the incubation period, the fragments are separated by length from longest to shortest. The resolution is so good that a difference of one nucleotide is enough to separate that strand from the next shorter and next longer strand. Each of the four dideoxynucleotides fluoresces a different color when illuminated by a laser beam and an automatic scanner provides a printout of the sequence.
Limitation : Limitations include non-specific binding of the primer to the DNA, affecting accurate read-out of the DNA sequence, and DNA secondary structures affecting the fidelity of the sequence
(c) Cellular totipotency : Totipotency is the ability of a single cell to divide and produce all the differentiated cells in an organism, including extraembryonic tissues and also the formation of a new organism. Totipotent cells formed during sexual and asexual reproduction include spores and zygotes.
Question 4.
(a) Briefly describe the essential components of the nutrient medium used for the plant tissue culture technique. Also, write the names of any two plant tissue culture media frequently used in the laboratory . [4]
(b) With reference to suspension culture, explain the following : [4]
(i) Achemostat.
(ii) A turbidostat.
(c) What are purines and pyrimidines ? Where are they located in a cell? [2]
Answer:
(a) Nutrient Medium : Virtually all tissue culture media are synthetic or chemically defined; only a few of.them use complex organics, e.g., potato extract, as their normal constituents. A synthetic medium consists of only chemically defined compounds. A variety of recipes have been developed since none of them is suitable for either all plant species or for every purpose. Most of these recipes have been elaborated from those of White (itself evolved from a medium for algae) and Gautheret (based on Knop’s salt solution) The composition is as follow :
Inorganic nutrients: In addition to C, H and O, all nutrient media provide the 12 elements essential for plant growth, viz., N, P, K, Ca, S, Mg (these six are called macronutrients, and are needed in concentrations >0.5 mmol L-1 or > 0.5 mM), Fe, Zn, Mn Cu, B and Mo (these six are known a micronutrients, and are required in concentrations <0.5 mmol L” ’ ) Generally, iron is provided as iron.EDTAcomplex to keep it available at higher (>5.8) pH. Nitrate is superior to ammonium as the sole N source, but use of NH+ checks the drift of pH towards alkalinity.
Vitamins : For optimum callus growth, the following vitamins are required . inositol, thiamine, pyridoxine and nicotinic acid of which thiamine is essential and the rest are promotory. Pantothenic acid is also known to be promotory but is not included in most of the recipes.
Carbon source : Sucrose (20-50 g L) is the most commonly used carbon source for all cultured plant materials, including even green shoots. In some systems, e.g., monocots, glucose may be superior to sucrose. Plant tissue can utilize other sugars like maltose, galactose, lactose, mannose and even starch but these are rarely used.
Growth regulator: The following growth regulators (GRs) are used in plant tissue culture. Auxins, e.g., IAA (indole-3-acetic acid), IBA (indole-3-butyric acid), NAA (napthalene acetic acid), NOA (naphthoxy acetic acid), 2, 4-D (2,4-dichlorophenoxy acetic acid) etc., are commonly used to support cell division and callus growth (especially 2. 4-D), somatic embryo (SE) induction, rooting, etc. Cytokinins like kinetin (furfurylamino purine), BAP (benzylamino purine), zeatin, 2-ip (isopentenyl adenine), TDZ (thidiazuron, a compound having cytokinin activity) are employed to promote cell division, regeneration of shoots often SE induction and to enhance proliferation and growth of axillary buds. Abscisic acid (ABA) promotes SE and shoot bud regeneration in many species are markedly improves SE maturation. Of the over 20 gibberellins known, GA3 is almost exclusively used. It promotes shoot elongation and SE germination.
Complex organic additives: In earlier studies, complex additives like yeast extract, coconut milk, casein hydrolysate, com milk, malt extract and tomato juice are used to support plant tissue growth. White’s medium, Murashige and Skoog (MS) are the two common media used for plant tissue culture.
(b) (i) Chemostat :A type of cell culture; a component of medium is in a growth limiting concentration; fresh medium is added at regular interv als and equal volume of culture is withdrawn. But in a chemostat, a chosen nutrient is kept in a concentration so that it is depleted very rapidly to become growth limiting, while other nutrients are still in concentrations higher than required. In such a situation, any addition of the growth-limiting nutrient is reflected in cell growth. Chemostats are ideal for the determination of effects of individual nutrients on cell growth and metabolism.
(ii) Turbidostat: A type of suspension culture; when culture reaches a predetermined cell density, a volume of culture is replaced by fresh medium; works well at growth rates close to the maximum. A continuous culturing method where the turbidity of the culture is kept constant by manipulating the rate at which medium is fed. If the turbidity falls, the feed rate is lowered so that growth can restore the turbidity to its start point. If the turbidity rise the feed rate is increased to dilute the turbidity back to its start point.
(c) Purines and pyrimidines are two of the basic units of the nucleic acids. They are found in a DNA and RNA in a cell Purines is a large sized double ring structure. It contains two bases, i.e., adenine and guanine, Pyrimidines are small sized, single ring structures. It contains three type of bases i.e., thymine, cytosine and uracil.
Question 5.
(a) Explain giving an example how recombinant DNA technology can be used for the formation of
the following: [4]
(i) A vaccine.
(ii) A plant with delayed fruit ripening.
(b) What is osmotic pressure ? Explain any one biochemical technique based on osmotic pressure. [4]
(c) What are dextro-rotatory and laevo-rotatory substances? [2]
Answer:
(a) (i) Recombinant vaccines : Vaccine is produced using recombinant DNA technology. A recombinant vaccine contains protein or a gene encoding a protein of a pathogen origin that is immunogenic A gene coding an immunogenic protein from the pathogen, is isolated, cloned and used for vaccine production The vaccines based on recombinant proteins are also called Sub-Unit Vaccines.
Whole protein vaccine : Hepatitis B vaccine is produced from surface antigens of transgenic yeast by r-DNA technology They can also be produced in genetically engineered microbes, cultured animal cells, possibly in insects and plants.
Recombinant – polypeptide vaccines : In some cases, the immunogenic portion of the protein- recombinant polypeptide is used as vaccine e.g., gene encoding B polypeptide (part of cholera enterotoxin – Ab A2 and B polypeptide) has been cloned and the recombinant B polypeptide produced is being used, in combination with inactivated cholera cells, as an oral vaccine in place of conventional injectable cholera vaccine. Immunogenicity of foot and mouth disease virus coat protein is due to its amino acids 114-160 and also 201 -213. They induce antibodies which neutralize the virus and thereby provide protection against the foot and mouth disease.
Live recombinant vaccine : The most advanced and promising approach in which concerned pathogen gene is introduced into the genome of selected viral / bacterial vector which is suitably attenuated and the live microorganisms are used for vaccination. Vaccinia virus appears to be more promising vector.
DNA vaccines: Recently vaccines based on pathogen naked DNA are being developed. Tue various approaches for DNA vaccines are as follow
- injection of pure DNA (or RNA) preparation into muscles
- reimplantation of autologus cells (cells of the individual to be vaccinated) into which the gene has been transferred and
- particle gun delivery of plasmid DNA which contains the gene in an expression casette e.g., skin cells They elicit humoral immune response and are usually shed off in a few days preventing long term persistence modified cells.
(ii) Delayed fruit ripening
A major problem in fruit marketing is the pre-mature ripening and softening dining transport of fruits. Consequently shelf-life of fruit remains short in the market. During ripening, genes encode the enzyme cellulase and polygalacturonase. Therefore, ripening process can be delayed by interfering the expression of these genes. In the U.S.A., a transgenic tomato named FlavrSavr (flavour saver) was produced where ripening is delayed by , lowering polygalacturonase activity.
A plant growth hormone ethylene is produced during fruit ripening and senescence. It is synthesised from S-adenosylmethionine through an intermediate compound 1-aminocyclopropane-l-carboxylic acid (ACC). There is a large number of bacteria that can degrade ACC. Therefore, bacterial gene (for ACC) deaminase associated with ACC degradation was isolated and introduced into tomato. In transgenic tomato, fruit ripening was delayed because it synthesised lower amount of ethylene (due to inhibition in ACC synthesis) than the normal tomatoes. Such tomatoes and other fruits can be transported to a longer distance without spoilage.
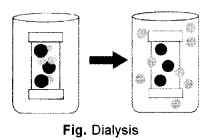
(b) The osmotic pressure is defined to be the pressure required to maintain an equilibrium, with no net movement of solvent. Osmotic pressure is a colligative property, meaning that the osmotic pressure depends on the molar concentration of the solute but not on its identity.
In biochemistry, dialysis is the process of separating molecules in solution by the difference in their rates of diffusion through a semi-permeable membrane, such as dialysis tubing. Typically a solution of several types of molecules is placed into a semi-permeable dialysis bag, such as a cellulose membrane with pores, and the bag is sealed. The sealed dialysis bag is placed in a container of a different solution, or pure water. Molecules small enough to pass through the tubing (often water, salt and other small molecules) tend to move into or out of the dialysis bag, in the direction of decreasing concentration. Larger molecules (often proteins, DNA, or polysaccharides) that have dimensions significantly greater than the pore diameter are retained inside the dialysis bag.
(c) Substances which rotate or deviate the plane of polarized light to the right (clockwise) are called dextrorotatory and are indicated by prefixing (+) or d to their names, e.g., d-glucose or (+) glyceraldehydes; Laevorotatory are the substances which rotate or deviate the plane polarized light to the left and are indicated by prefixing L or (–) to their names; e.g., L-alanine or (–) glyceraldehydes.
Question 6.
(a) Write short notes on : [4]
(i) Single nucleotide polymorphism.
(ii) Bioinformatics databases.
(b) How are biomolecules separated by the following techniques: [4]
(i) Chromatography.
() Centrifugation.
(c) Give two differences between enzymes and inorganic catalysts: [2]
Answer:
(a) (i) Single nucleotide polymorphisms, commonly called SNPs (pronounced “snips”), are the most common type of genetic variation in a nucleotide sequence due to change even in a single base among different individuals. Thus, each SNP represents a difference in a single DNA building block, called a nucleotide. For example, a SNP may replace the nucleotide cytosine (C) with the nucleotide thymine (T) in a certain stretch of DNA. In human genome, SNPs occur at 1.6-3.2 million site. Due to the changes in bases SNPs affect the gene function. DNA fingerprinting of individuals is possible due to these genetic variations in non-coding parts of genome.
(ii) Database : A database is an organized collection of data for one or more multiple uses. One way of classify ing databases involves the type of content, for example : bibliographic, full-text, numeric and image.
Biological databases are libraries of life sciences information, collected from scientific experiments, published literature, high-throughput experiment technolog}, and computational analysis. They contain information from research areas including genomics, proteomics, metabolomics, microarray gene expression, and phylogenetics. Information contained in biological databases includes gene function, structure, localization (both cellular and chromosomal), clinical effects of mutations as well as similarities of biological sequences and structures.
Examples:
- ENA (European Nucleotide Archive) – primary nucleotide data, incorporating EMBL- Bank.
- UniProt-protein databases.
- PDB (Protein Data Bank)-biological macromolecular structure.
(b) (i) Chromatography is a technique of separation of biomolecules involves a sampling mixture containing biomolecules being dissolved in a mobile phase (which may be a gas, a liquid or a supercritical fluid) due to their differential adsorption over an adsorbent medium. The mobile phase is then forced through an immobile, immiscible stationary phase. The phases are chosen such that components of the sample mixture have differing solubilities in each phase.
A component which is quite soluble in the stationary phase will take longer to travel through it than a component which is not very’ soluble in the stationary phase but very soluble in the mobile phase. As a result of these differences in mobilities, sample mixture components will become separated from each other as they travel through the stationary’ phase.
Techniques such as H.P.L.C (High Performance Liquid Chromatography) and GC. (Gas Chromatography) use columns—narrow tubes packed with stationary phase, through which the mobile phase is forced. The sample is transported through the column by continuous addition of mobile phase. This process is called elution. The average rate at which an analyte moves through the column is determined by the time it spends in the mobile phase.
(ii) Centrifugation is a process that involves the use of the centrifugal force for the sedimentation of the components of a mixture with a centrifuge. More-dense components of the mixture migrate away from the axis of the centrifuge, while less-dense components of the mixture migrate towards the axis. Chemists and biologists may increase the effective gravitational force on a test tube so as to move rapidly and completely cause the precipitate (‘’pellet”) to gather on the bottom of the tube. The remaining solution is properly called the “supernate” or ‘ supernatant liquid”. The supernatant liquid is then either quickly decanted from the tube without disturbing the precipitate, or withdrawn with a Pasteur pipette. For example, microcentrifuges are used to process small volumes of biological molecules, cells, or nuclei.
(c) Differences between enzyme and inorganic catalyst are mentioned as follows :
| Enzymes | Catalysts |
| (i) Enzymes are complex organic proteins. | (i) Catalysts are simple inorganic molecules. |
| (ii) Enzymes catalyses specific types of reactions. | (ii) Catalysts have a wide range. |
Question 7.
(a) Differentiate between : [4]
(i) Prokaryotic genome and Eukaryotic genome.
(ii) Somatic embryo and Zygotic embryo.
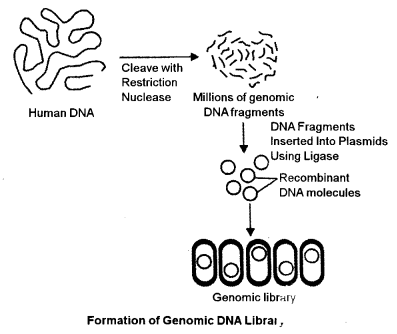
(b) Explain how a genomic DNA library is formed. How does it differ from cDNA library ? [4]
(c) Name any two inborn metabolic disorders in human beings. Also, write one main symptom for each of them.
Answer:
(a) (i) Prokaryotic Genome:
- Genome are much smaller and simpler.
- Highly repetitive DNA is not found.
- It is without a limiting membrane.
- It is a naked double strand of DNA.
Eukarvotic Geflorne:
- Genome are larger and complex.
- Occurrence of highly repetitive DNA is found.
- It is bounded by nuclear membrane.
- Double strand of DNA is associated with histone proteins.
(ii) Somatic Embryo: Somatic embryos are formed from plant cells other than egg that are not normally involved in the development of embry os, e.g., callus or explant. No endosperm or seed coat is formed around a somatic embryo.
Zygotic Embryo: Zygotic embryo is formed as a result of double fertilization of the ovule, giving rise to two distinct structures; the zygote and primary endosperm nucleus giving rise to the plant embryo and the endosperm which together go on to develop into a seed.
(b) Construction of Genomic Library : The process of subdividing genomic DNA into clonable elements and inserting them into host cells is called creating a library.
A complete library, by definition, contains the entire genomic DNA of the source organism and is called as genomic library. A genomic library is a set of cloned fragments of genomic DNA. The process of creating a genomic library includes four steps :
In the first step the high molecular weight genomic DNA is separated and subjected to restriction enzy me digestion by using two compatible restriction enzymes.
In the second step, the fragments are then fractionated or separated by using agarose gel electrophoresis to obtain fragments of required size.
These fragments are then subjected to alkaline phosphatase treatment to remove the phosphate. In the third step, the dephosphorylated insert is ligated into vector which could be a plasmid, phage or cosmid, depending upon the interest of the researcher.
In the last step, the recombinant vector is introduced into the host by electroporation and amplified in host. In principle, all the DNA from the source organism is inserted into the host but this is not fully possible as some DNA sequences escape the cloning procedure. Genomic library is a source of genes and DNA sequences. A genomic library is a set of cloned fragments of genomic DNA. Prior information about the genome is not required for library construction for most organisms. In principle, the genomic DNA, after the isolation, is subjected to RE enzyme for digestion to
generate inserts.
cDNA libraries V/S Genomic libraries:
- Genomic library is a mixture of fragments of genomic DNA while cDNA obtained from mRNA may cloned to give rise to a cDNA library.Genomic library’ contains DNA fragments that represent genes as well as those that are not genes. In contrast cDNA library contains only those genes that are expressed in the concerned tissue/organism. In both cases, a mixture of fragments is used for cloning to establish the library.
- Use of cDNA is absolutely essential when the expression of an eukaryotic gene is required in a prokaryote.
- Eukaryotic cDNAs are free from intron sequences.
- As a result of the above, they are smaller in size than the corresponding genes, i.e., the genes that encoded them.
- A comparison of the cDNA sequence with the corresponding genome sequence permits the delineation of intron/exon boundaries.
- The contents of cDNA libraries from a single organism will vary widely depending on the developmental stage and the cell type used for preparation of the library. In contrast, the genomic libraries will remain essentially the same irrespective of the developmental stage and the cell type used.
- A cDNA library will be enriched for abundant mRNAs, but may contain only a few or no clones representing rare mRNAs.
(c) Alkaptonuria : This was one of the first metabolic diseases described by Garrod in 1908. It is an inherited metabolic disorder produced due to deficiency of an oxidase enzyme requiredfor breakdown ofhomogentisic acid (also called alcapton, hence, alkaptonuria is also written as alcaptonuria). Lack of the enzyme is due to the absence of the normal form of gene that controls the synthesis of the enzyme. Hence, homogentisic acid then accumulates in the tissues and is also excreted in the urine. The most commonly affected tissues are cartilages, capsules of joints, ligaments and tendons. The urine of these patients if allowed to stand for some hours in air, turns black due to oxidation of homogentisic acid.
Phenylketonuria (PKU; Foiling. 1934): It is an inborn metabolic disorder in which the homozygous recessive individual lacks the enzyme phenylalanine hydroxylase needed to change phenylalanine (amino acid) to tyrosine (amino acid). Thus, the biochemical abnormality in PKU is an inability to convert phenylalanine into tyrosine leading to hyperphenylalaninemia. Lack of the enzyme is due to the abnormal autosomal recessive gene on chromosome 12. This defective gene is due to substitution. Affected babies are normal at birth but within a few weeks there is rise (30-50 times) in plasma phenylalanine level which impairs brain development. Usually by six months of life severe mental retardation becomes evident.
If these children are not treated about one-third of these children are unable to walk and two-thirds cannot talk. Other symptoms are mental retardation, decreased pigmentation of hair and skin and eczema. Although large amounts of phenylalanine and its metabolites are excreted in the urine and sweat, yet it is believed that excess phenylalanine or its metabolites contribute to the brain damage in PKU. The heterozygous individuals are normal but carriers. It occurs in about 1 in 18000 births among white Europeans. It is very rare in other races.
Question 8.
(a) Give the step-wise procedure of Southern blotting technique. Mention any two important applications of this technique. [4]
(b) What are blunt ends and sticky ends ? How are they formed ? [4]
(c) Name any two industrial enzymes and give their uses. [2]
Answer:
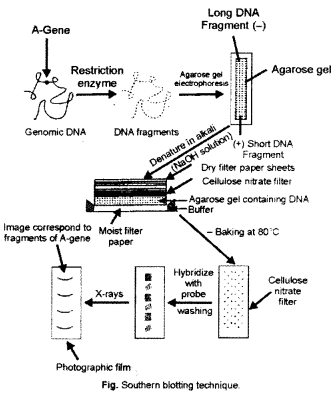
(a) Southern blotting technique : In 1975, Edward M. Southern developed technique of DNA separation and its hybridization. Therefore, in his honor this technique is known as ‘ Southern blotting or Southern hybridization technique’. A specific DNA fragment can be separated and identified in a heterologous population of DNA molecules on the basis of binding of DNA probe with its complementary DNA strand.
The genomic DNA is isolated from the clone and digested with restriction enzymes. The DNA fragments are separated by agarose gel electrophoresis (Fig.). Different DNA bands are formed on agarose gel which represent DNA fragments of varying sizes. These fragments are transferred from gel to nylon or nitrocellulose membrane. The process of DNA transfer is called ‘blotting’.
A nitrocellulose membrane is put over the gel. Many layers of filter paper are placed over nitrocellulose membrane. This assembly is put in a container having NaOH solution. NaOH denatures DNA and results in formation of single stranded DNA. DNA fragments are transferred from gel to membrane by capillary action.
In addition, DNA fragments can also be transferred by vacuum blotting and centrifugation. The DNA fragments are fixed to membrane by using UV radiation or baking at 80°C. The pattern of DNA bands on membrane corresponds to the position of DNA on gel. The membrane is put in solution containing radio labelled DNA probe and incubated for some time. DNA probe hybridizes complementary DNA fragments fixed on membrane. It is gently washed at 12°C and dried.
The membrane is exposed through a photographic film. DNA bands formed on photographic film corresponds to the original position of DNA fragments present on agarose gel.
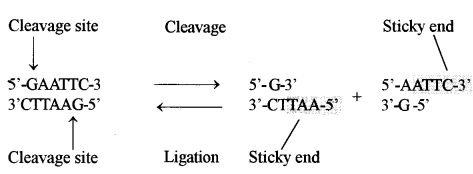
(b) Restriction enzymes are bacterial proteins that have the ability, to cut both the strands of the DNA molecule at a specific nucleotide sequence. There are hundreds of these restriction enzymes and each can cut the DNA at a specific point and the resulting DNA fragments are all of different lengths. Restriction enzymes can either produce sticky ends or blunt ends.
EcoRI enzyme binds to a region of having specific palindromic’ sequence (where two strands are identical when both are read in the same polarity i.e., in 5’→ 3’ direction). The length of this region is 6 base pairs i.e., hexanucleotide palindrome. It cuts between G and A residues of each strand and produces two single stranded complementary cut ends which are asymmetrical having 5’ overhangs of 4 nucleotides. These ends are called sticky ends or cohesive ends. Because nucleotide bases of this region can pair and stick the DNA fragments again as given below :}
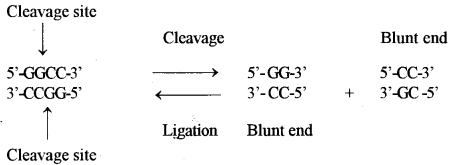
On the other hand, there are some other Type II restriction enzymes which cleave both strands of DNA at the same base pairs but in the centre of recognition sequence, and results in DNA fragments with blunt ends or flush ends. For example Hae111 (isolated from Haemophilus aegypticus, the order of enzyme III), four nucleotide long palindromic sequence and cuts symmetrically the both DNA strands and forms blunt ends as below:
(c) Alpha Amylase : It is widely used enzyme in Food industry and in Laundry detergent.
Papain : It is used in Medicine, Food and Textile industries.
Question 9.
(a) Enlist the main steps in the regeneration of a complete plant from an explant. [4]
(b) Given below is a list of four bio molecules found in a living cell. For each of them, write the class of biomolecules they belong to and their location in a living cell: [4]
(i) Histones.
(iii) Haemoglobin.
(ii) mRNA.
(iv) Glycogen.
(c) Write any two uses of transgenic plants. [2]
Answer:
(a) Steps involved in in vitro regeneration of a complete plants. Regeneration refers to the development of organised structures like roots, shoots, flower buds, somatic embryos (SEs), etc. from cultured cells/tissues; the term organogenesis is also used to describe these events. Root regeneration occurs quite frequently but it is useful only in case of shoots and embryo germination. Only shoot and SE regenerations give rise to complete plants which is essential for applications of tissue culture technology in agriculture and horticulture. Regeneration may occur either directly from the explant or may follow an intervening callus phase.
Basic technique of Plant Tissue Culture: The basic technique of plant tissue culture involves the following steps:
- Preparation and sterilisation of suitable nutrient medium : Suitable nutrient medium as per objective of culture is prepared and transferred into suitable containers. Culture- medium is rich in sucrose, minerals, vitamins, and hormones. Yeast extract, coconut milk are also added. The culture is completely sterilized in an autoclave.
- Selection of explants : Selection of explants such as shoot tip should be done.
- Sterilisation of explants: Surface sterilisation of the explants by disinfectants (e.g., sodium hypochlorite or mercuric chloride) and then washing the explants with sterile distilled water is essential.
- Inoculation: Inoculation (transfer) of the explants into the suitable nutrient medium (which is sterilised by autoclaving to avoid microbial contamination) in culture vessels under sterile conditions is done.
- Incubations : Growing the culture in the growth chamber or plant tissue culture room, having the appropriate physical condition (i.e., artificial light: 16 hours of photoperiod), temperature (- 26°C) and relative humidity (50 – 60%) is required.
- Regeneration : An unorganized mass of cells developing from explants is called callus. The callus gives rise to embryoids which can develop into whole plant if the medium is provided with proper concentration of hormones. This property’ of developing every somatic cell into a Ml fledged plant is called totipotenev. Regeneration of plant, from cultured plant tissues is carried out.
- Hardening: Hardening is gradual exposure of plantlets to an environmental conditions.
- Plantlet transfer : After hardening, plantlets are transferred to the greenhouse or field conditions following acclimatization (hardening) of regenerated plants.
