ISC Biotechnology Previous Year Question Paper 2011 Solved for Class 12
Maximum Marks: 80
Time allowed: Three hours
- Candidates are allowed additional 15 minutes for only reading the paper. They must NOT start writing during this time.
- Answer Question 1 (Compulsory) from Part I and five questions from Part II, choosing two questions from Section A, two questions from Section B and one question from either Section A or Section B.
- The intended marks for questions or parts of questions are given in brackets [ ].
- Transactions should be recorded in the answer book.
- All calculations should be shown clearly.
- All working, including rough work, should be done on the same page as, and adjacent to the rest of the answer.
Part-I
(Compulsory)
Question 1
(a) Mention any one significant difference between each of the following : [5]
(i) Nucleotide and nucleoside.
(ii) Plasmids and phages.
(iii) Fluorescence spectrometry and mass spectrometry.
(iv) Primer and primases.
(v) Introns and exons.
(b) Answer the following questions : [5]
(i) What are oil eating bacteria?
(ii) What are single cell proteins?
(iii) Who proposed the Operon concept of gene regulation?
(iv) Explain the term transposons.
(v) Name one polysaccharide found in the cell wall of bacteria and one polysaccharide found in fungi.
(c) Write the full form of the following: [5]
(i) NCBI
(ii) EST
(iii) ROM
(iv) PACE
(v) FMN
(d) Explain briefly: [5]
(i) RNA dependent DNA polymerase
(ii) EMBL
(iii) Isoelectric focussing
(iv) Cosmids
(v) Vitamins
Answer:
(a) (i) Nucleotide: The nucleotide is a condensation product of heterocyclic nitrogen base, a pentose sugar like ribose or deoxyribose, and a phosphate or polyphosphate group.
Nucleoside: Nucleoside consisting of a pentose sugar, usually ribose or deoxyribose, and a nitrogen base-purine or pyrimidine.
(ii) Plasmids: Plasmids are the extra-chromosomal, independent, self replicating, circular, double stranded DNA molecules naturally found in all bacteria and some fungi.
Phages: Bacteriophage (i.e., viruses that infect bacteria) are obligate intracellular parasites that multiply inside bacteria by making use of some or all of the host biosynthetic machinery.
(iii) Fluorescence Spectrometry: Fluorescence spectrophotometry is a technique that assay the state of a biological system by studying the absorption spectra of different radiations and its interactions with fluorescent probe molecules.
Mass Spectrometry: Mass spectrometry (MS) is an analytical technique that involves separation of ions of different mass and energy when fixed magnetic and electric fields are employed.
(iv) Primer: A primer is a short strand of RNA that serves as a starting point for DNA synthesis They are required for DNA replication because DNA polymerases can only add new nucleotides to an existing strand of DNA.
Primases: Primase is an enzyme that catalyzes the synthesis of a short RNA segment (called a primer) complementary to a ssDNA template.
(v) Introns: Introns are non-coding DNA base sequences , which are found between exons, but are not transcribed part of mature mRNA.
Exons: Exons are coding DNA base sequences that are transcribed into wzRNA and finally code for amino acids in the proteins.
(b) (i) Bacteria that decompose and use oil as their source of energy are called as oil eating bacteria e.g., Pseudomonasputida, P.capacia etc. are efficient degraders created through genetic engineering. They need to be established in environment. They (“the oil-eating bacteria”) could digest about two- thirds of the hydrocarbons that would be found in a typical oil spill.
(ii) The term single cell protein (SCP) refers to biomass of dry cells of microorganisms such as yeast, bacteria, mushroom (fungi) and algae which grow on different carbon sources. The name “single cell protein” was used for the first time by the M.I.T. Professor Carol Wilson.
(iii) Jacob and Monod proposed the Operon concept of Gene Expression in bacteria.
(iv) Transposons : Transposons are sequences of DNA that can move around to different positions within the genome of a single cell. Transposons are called “Jumping genes”, and are examples of mobile genetic elements.
Two types of Transposons are possible :
- Retrotrahsposons and
- DNA transposons.
In this process, they may :
- cause mutations
- increase (or decrease) the amount of DNA in the genome.
(v) Peptidoglycans (mucopeptides, glycopeptides, mureins) are the structural elements of bacterial cell walls. Fungi possess cell walls made-up of chitin, polymer of glucosamine,
(c) (i) NCBI: National Centre for Biotechnology Information
(ii) EST : Expressed sequenced tag
(iii) ROM : Read only memory
(iv) PAGE : Polyacrylamide gel electrophoresis
(v) FMN : Flavin mononucleotides.
(d) (i) RNA-dependent DNA polymerase enzyme is used for the synthesis of a complementary DNA strand from RNA molecule as a template. It is produced by HTV and other retroviruses which help them to synthesize DNA from their viral RNA.
(iii) Isoelectric focusing (IEF), also known as electrofocusing, is a technique for separating different molecules by their electric charge differences. It is a type of zone electrophoresis, usually performed on proteins in a gel.
(iv) Cosmids are constructed by uniting some part of the bacterial plasmids with a ‘ori’ gene, an antibiotic selection marker and a cloning site with one or more recently two ‘cos’ sites derived from bacteriophage lambda.
(v) A vitamin is an organic compound required as a nutrient in very small amounts by an organism. They act as co-factor of an enzyme. It cannot be synthesized in sufficient quantities by an organism, and must be obtained from the diet.
Part-II
(Answer any five questions)
Question 2.
(a) Mention two important chemical properties of each of the following: [4]
(i) Monosaccharides
(ii) Proteins .
(b) What are restriction enzymes ? How do thev act ? Give any two examples of restriction enzymes. [4]
(c) What are derived lipids ? Give an example. [2]
Answer:
(a) (i) Chemical properties of monosaccharides :
When a & p isomeric forms of D-glucose are dissolved in water. Their optical rotation changes with time and approaches the final equilibrium value. +53°. This change is known as mutarotation. Mutarotation occurs because of slow conversion of α -D glucose and β-D glucose via open chain form until equilibrium is established giving a constant rotation +53°.
Acetals traditionally derive from the product of the reaction of an aldehyde with excess of alcohol, whereas the name ketal derives from the product of the reaction of a ketone with excess alcohol.
(ii) Chemical properties of proteins :
- Proteins when hydrolyzed by acidic agents, like cone. HC1 yield amino acids in the form of their hydrochlorides.
- Sanger’s reaction : Proteins react with FDNB reagent to produce yellow coloured derivative, DNB amino acid.
- Xanthoproteic test: On boiling proteins with cone. HN03, yellow colour develops due to presence of benzene ring.
- Folin’s test: This is a specific test for tyrosine amino acid, where blue colour develops with phosphomolybdotungstic acid in alkaline solution due to presence of phenol group.
(b) The restriction enzymes are called ‘molecular scissors’. Restriction enzymes are DNA-cutting enzymes present in bacteria. They are obtained from them for use in genetic enginccring-rDNA technology. As sequence are cut within the DNA molecule, they are often called restriction endonucleases.
A restriction enzyme recognizes and cuts DNA only at a particular sequence of nucleotides. For example, the bacterium Hemophilus aegypticus produces an enzyme named Hae III that cuts DNA whenever it identifies the recognition sequence.
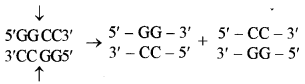
A cut is made between the adjacent G and C. HaeIII cleaves both the strands of DNA at the same base pairs producing “blunt” ends.
Examples : EcoRI, HindIII and Bam HI.
(c) Derived lipids are either lipid-like chemicals (e.g., sterols) or derivatives of lipids e.g., terpenes. Prostaglandis and Choline. They are derived from simple and compound lipids by hydrolysis.
Question 3.
(a) Give an account of various enzymes that play a role in the process of DNA replication. [4]
(b) Write short notes on : [4]
(i) Secondary structure of proteins
(ii) Designer oils.
(c) What is meant by synchronization of cell culture. [2]
Answer:
(a) Topoisomerase : Cause single strand breaks and religation.
Helicase: Unwounds a portion of the DNA Double Helix.
RNA Primase : Attaches RNA primers to the replicating strands.
DNA Polymerase delta (δ) : Binds to the 5′ – 3′ strand in order to bring nucleotides and synthesise the daughter leading strand.
DNA Polymerase epsilon (a): Binds to the 3′ – 5′ strand in order to synthesise discontinuous segments starting from different RNA primers.
Exonuclease (DNA Polymerase I): Identifies and removes the RNA Primers.
DNA Ligase : Adds phosphate in the remaining gaps of the phosphate – sugar backbone.
Nucleases : Remove wrong nucleotides from the daughter strand.
(b) (i) Secondary Structure (2° structure): It is development of new stearic relationships amongst the amino acids for protecting their peptide bonds through formation of intrapolypeptide and interpolypeptide hydrogen bonds. Secondary structure is of three types — α-helix, β-pleated and collagen helix. The prefixes α and β signify the first and second types of secondary structure discovered by Pauling and Corey (1951).
α-Helix : The polypeptide chain is spirally coiled, generally in a clockwise or right¬handed fashion (Fig ). There are 3.6 amino acid residues per turn of the spiral. The . spiral is stabilized by straight hydrogen bonds between imide group (-NH-) of one amino acid and carbonyl group (-CO-) of fourth amino acid residue. In this way all the imide and carbonyl groups become hydrogen bonded. R-groups occur towards the outer side of a-helix. a-helix is the final structure in certain fibrous proteins, e.g., keratin (hair, nail, horn), epidermin (skin).
β-Pleated Sheets : Two or more polypeptide chains come together and form a sheet. Condensation is little. However, twisting does occur. The same polypeptide may fold over itself to form two strands for β-pleating. Adjacent polypeptide chains may occur in parallel (e.g., p-keratin) or antiparallel (e.g., silk fibroin). Straight hydrogen bonds occur between imide (-NH-) group of one polypeptide and carbonyl (-CO-) group of adjacent polypeptide. Cross-linkages help in stabilisation of p-pleated sheets.
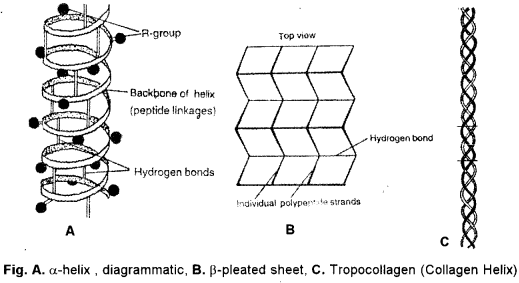
Collagen Helix : Collagen has a large amount of glycine (25%) and proline (and hydroxyprohne, 25%). It cannot form a-helix due to them. Three of its polypeptide each having about 1000 amino acid residues, come together with each forming an extended left-handed helix. They run parallel, form a right-handed super-helix that is stabilised by hydrogen bonds amongst the three. The triple helix of collagen is often called tropo-collagen. Its one end is stabilised by -S-S- linkages amongst the three chains. Collagen occurs in those tissues where extensibility is limited, e.g., connective tissue, tendons, bones.
(ii) Designer Oil: “Designer oif’ that reduces LDL (“bad”) blood cholesterol levels in humans and increases energy expenditure, which may prevent people from gaining weight. The oil incorporates a phytosterol-based functional food ingredient Phytrol (TM) from Forbes into oil using proprietary technology.
(c) Cell culture synchronisation : Cells in suspension cultures vary greatly in size, shape, DNA, and nuclear content. Moreover, the cell cycle time varies considerably within individual cells. Therefore, cell cultures are mostly asynchronous. It is essential to manipulate the growth conditions of an asynchronous culture in order to achieve a higher degree of synchronisation. A synchronous culture is one in which the majority of cells proceed through each cell cycle phase (Gl, S, G2 and M) simultaneously. Synchronisation can be achieved by following methods.
- Physical methods include selection by volume (size of cell aggregate.)
- Chemical methods include starvation (depriving suspension cultures of an essential growth compound and culture supplying).
- Chemical methods include inhibition (temporarily blocking the progression of events in the cell cycle using a biochemical inhibitor and then releasing the block).
Question 4.
(a) Write the steps in the process of activation of amino acids during the process of translation. [4]
(b) What is the significance of genetic code in protein synthesis ? Mention four important characteristics of the genetic code. [4]
(c) Name the important steps in a single cycle of polymerase chain reaction. [2]
Answer:
(a) The adapter function of the tRNA molecules require the charging of each specific tRNA with its specific amino acid. Since there is no affinity of nucleic acids for specific functional groups of amino acids, this recognition must be carried out by a protein molecule capable of recognizing both a specific tRNA molecule and a specific amino acid.
Activating enzymes called aminoacyl-tRNA synthetases couple each amino acid to its appropriate set of tRNA molecules. There are 20 synthetases for each of the 20 natural amino acids. The reaction involves two steps :
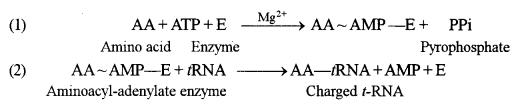
The amino acid-tRNA bond is a high-energy linkage that easily reacts with the amino group of the next amino acid in a protein sequence to form a peptide bond during protein synthesis.
(b) Messenger RNA contains a triplet sequence of nitrogen bases which code for the specific amino acids, used to make polypeptide chains. Each of the sets of three bases is known as a codon or genetic code.
Translation starts with a chain initiation codon (start codon). The most common start codon is AUG which is read as methionine or, in bacteria, as formyl methionine. The three stop codons have been given names : UAG is amber, UGA is opal, sometimes also called umber and UAA is ochre. Stop codons are also called “termination” or “non-sense” codons.
Characteristics of Genetic Code
- Triplet code : Three adjacent nitrogen bases constitute a codon which specifies the placement of one amino acid in a polypeptide.
- Start signal: Polypeptide synthesis is signaled by AUG or methionine codon and GUG — Valine codon. They have dual function.
- Stop signal: Polypeptide chain termination is signaled by three termination codons — UAA, UAG and UGA. They do not specify any amino acid and are hence also called non-sense codon.
- Universal code : The genetic code is applicable universally i.e., the codon specifies the same amino acid from a virus to a tree or human being.
- Non-ambiguous codon : One codon specifies only one amino acid and not any other.
(c) PCR Stages:
- Denaturation step
- Primer Annealing step
- Extension/elongation step (Polymerization)
Question 5.
(a) Write notes on : [4]
(i) Proteomics.
(ii) De-differentiation and Re-differentiation.
(b) Mention the objectives of germplasm conservation using the cell culture technique. What are the limitations of conservation of germplasm using conventional methods? [4]
(c) The base sequence of one strand of DNA is 3′ CATGAC 5′. What will be the base sequence of its : [2]
(i) Complementary DNA strand.
(ii) Complementary RNA strand.
Answer:
(a) (i) Proteomics is the identification, analysis and large scale characterisation of proteome (i.e., the total protein components) expressed by any given cells, tissues and organs under the defined conditions. The major objectives of proteomics are
- to characterise post- transcriptional modifications in protein, and
- to prepare 3D map of cell indicating the exact location of protein.
Proteomics is the direct outcome of advancement made for nucleotide sequencing of different genomes in large scale. This helps to identify various proteins. Generation of information about protein is necessary. Because, protein governs the phenotypic characters of the cells. Merely genome study cannot provide the understanding of mechanism of disease development and various developmental changes occurring in organisms including humans. Moreover, target drugs for many kinds of diseases can be prepared only after understanding the protein modification and protein functions. There are many areas of modem proteomics such as protein expression, protein structure, protein localisation, protein-protein interaction, etc.
(ii) Dedifferentiation: It is a cellular process which occurs in lower life forms such as worms and amphibians in which a partially or terminally differentiated cell reverts to an earlier developmental stage.
Redifferentiation : It is a process by which a group of once differentiated cells return to their original specialised form.
(b) Germplasm Conservation:
The sum total of all the genes present in a crop and its related species constitutes its germplasm; it is ordinarily represented by a collection of various strains and species. Germplasm provides the raw materials (= genes), which the breeder uses to develop commercial crop varieties. Therefore, germplasm is the basic indispensable ingredient of all breeding programmes, and a great emphasis is placed on collection, evaluation and conservation of germplasm.
Limitations of conventional methods:
- Conventionally, germplasm is conserved as seeds stored at ambient temperature, low temperature or ultralow temperature. But many crops produce recalcitrant or short-lived seeds, and in case of clonal crops seeds are not the best material to conserve in view of their genetic heterogeneity and unknown worth.
- Roots and tubers loose viability rapidly and their storage requires large space, low temperature and is expensiv e.
- In addition, materials modified by genetic engineering may sometimes be unstable, and hence may need to be conserved intact for future use.
In such cases, the following approaches of germplasm conservation may be applied:
- freeze preservation,
- slow-growth cultures.
- DNA-clones and
- desiccated somatic embryos/artificial seeds.
(c) DNA -3′ CATGAC 5′
- COMPLIMENTARY DNA: 5′ GTACTG 3′
- COMPLIMENTARY RNA: 5′ GUACUG 3′
Question 6.
(a) Why are enzymes called biocatalysts ? Briefly discuss the mode of enzyme action. [4]
(b) Explain the principle and applications of each of the following biochemical techniques: [4]
(i) Gel permeation.
(ii) Colorimetry.
(c) How are the diseases cystic fibrosis and albinism caused? [2]
Answer:
(a) Catalysts are chemical substances used to speed up chemical reaction without any change of its own. In biological systems, certain biomolecules known as enzymes act in the same manner i.e., they mediate different biochemical reactions but themselves remain unaltered so they are termed as biocatalysts.
Mode of enzyme action : It could be explained by following models :
(b) (ii) Colorimetry
Principle : Colorimetry is the technique of determining the concentration of a chemical in a solution, if it has a colour, is to measure the intensity of the colour and relate the intensity of the colour to the concentration of the solution. In colorimetry, two fundamental laws are applied :
- firstly , a Lambert’s law. relates the amount of light absorbed and the distance it travels through an absorbing medium; and
- Secondly, Beer’s law relates light absorption and the concentration of the absorbing substance.
Application : It is used extensively for identification and determination of concentrations of substances that absorb light (any colour solution). A simple application lies in comparing intensities of radiation transmitted through layers of different thicknesses of two solutions of the same absorbing substance, one with a known concentration, the other unknown.
(c) Cystic fibrosis: A disease that results from a decrease in fluid and salt secretion by a transport protein known as Cystic Fibrosis Transmembrane Regulator (CFTR). As a result of this defect secretion from the pancreas is blocked and heavy dehydrated mucus accumulates in the lungs leading to chronic lung infection.
Albinism: It is a recessive disorder due to non-conversion of typosine into melanin because of gene mutation.
Albinism are of two types :
- Type 1 albinism is caused by defects that affect production of the pigment, melanin.
- Type 2 albinism is due to a defect in the ‘P” gene. People with this type have slight coloring at birth.
Question 7.
(a) Explain how ‘Dolly’ was created. What is its importance in the field of biotechnology? [4]
(b) Differentiate between: [4]
(i) PCR and Gene Cloning.
(ii) Batch Culture and Continuous Culture.
(c) What are polysaccharides ? How are they formed? [2]
Answer:
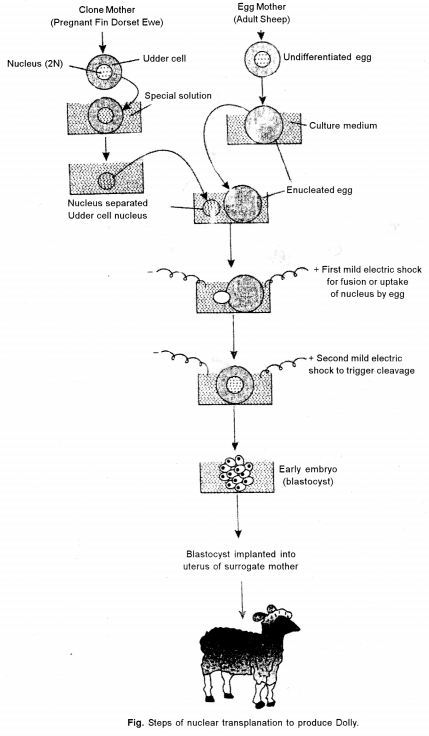
(a) Dolly, was the first mammal to be cloned by Wilmut et. al (1977). Dolly was the end result of a long research program founded by the British government at the Roslin Institute in Scotland. They used the technique of somatic cell nuclear transfer, where the cell nucleus from an adult udder cell is transferred into an unfertilized oocyte that has had its nucleus removed. The hybrid cell is then stimulated to divide by an electric shock, and the blastocyst that is eventually produced is implanted in a surrogate mother. Dolly was the first clone produced from a cell taken from an adult animal.
The following steps are involved in creation of Dolly :
Isolate donor nucleus : Isolate the nucleus from a somatic (non-reproductive) cell of a udder of adult donor sheep. The nucleus contains all the complete genetic material of the organism. A very small needle and syringe (suction device) is used to penetrate through the cell membrane to hold the nucleus and remove it from the cell.
Get unfertilized eggs: Retrieve some unfertilized egg cells (reproductive) from a female sheep. Many eggs are needed since not all of them will survive the various steps of cloning.
Remove the egg’s nucleus: Remove the egg cell’s nucleus, which contains only one-half of the sheep’s genetic material.
Insert donor nucleus : Insert the nucleus, with all its complete genetic material, isolated from the donor sheep mammal in Step 1 into the cytoplasm of egg cell that has no nuclear material. The egg’s genetic material now contains all traits from the donor adult. This egg is genetically identical to the donor adult cells.
Transfer the egg into womb : Transfer the egg into a receptive female sheep’s womb. Those eggs that survive and implant will continue to develop into embryos. When the offspring is born, it is a clone (genetically identical) of the donor sheep. After cloning was successfully demonstrated through the production of Dolly, many other large mammals have been cloned, including horses and bulls.
(b) (i) PCR:
- Amplification of desired segment of DNA within hours.
- No host cell required.
Gene cloning:
- Amplification of a desirable gene takes more time under lab condition.
- Host cell is required.
(ii) Batch culture:
- In this culture, the same medium and all the cells produced are retained in the culture vessel. Fresh medium is not added.
- The cell number of biomass exhibits a typical sigmoid curve.
Continuous culture:
- In this culture, both cells and used medium are taken out from the continuous culture and replaced by equal volume of fresh medium.
- Here, the cell population is a maintained in a steady state by regularly replacing a portion of used or spent medium.
(c) Polysaccharides are complex polymeric carbohydrate structures, formed by polymerization of monomeric repeating units (either mono-or di-saccharides) joined together by glycosidic bonds.
Example: starch, glycogen, etc. Polysaccharides are formed by enzyme catalyzed condensation reaction between any number of saccharide units.
Starch : It is the nutritional reservoir of plants. They are formed by the polymerisation/condensation of glucose molecules. Two types of starch are possible unbranched starch known as Amylase and branched starch known as Amylopectin. They actually differ in glycosidic linkage.
Glycogen : It is the storage form of glucose in animals. Two chains of glucose molecules joined by an alpha-1, 4-glycosidic bonds are linked by an alpha-1. 6-glycosidic bond to create a branch point.
Question 8.
(a) Briefly explain the concept and application of: [4]
(i) In vitro pollination.
(ii) Protoplast fusion.
(b) Mention the significance of plants obtained by each of the follow ing types of plant tissue culture techniques : [4]
(i) Endosperm culture.
(ii) Anther culture.
(c) State the main achievements of the Human Genome Project. [2]
Answer:
(a) (i) Pollination and fertilization under in vitro conditions offer an opportunity for producing hybrid embry os among plants that cannot be crossed by conventional methods of plant breeding. In nature, Intergeneric or Interspecific hybridization occurs very rarely. This is due-to barriers hindering the growth of the pollen tube on the stigma or style. In such cases, the style or part of it can be excised and pollen grains either placed on the cut surface of ovary or transferred through a hole in the wall of ovary. This technique, called intraovarian pollination, has been successfully applied in such species as Papaver somniferum, Eschscholtiza California, Argemone mexicana.
Ovular Pollination : In this in vitro pollination method, pollen tubes directly penetrate isolated ovules. Interspecific crossing barriers do not occur during penetration of the pollen tubes into the micropyle. Completion of pollen tube penetration is much earlier than the division of the generative cell in sperm cells which may be the primary cause for the failure in fertilization.
Placental Pollination : In this in vitro pollination method, the ovaries are cut into sectors on the day or after the day of stigma receptivity. Each sector contains a placenta with a row of ovules without or with ovary wall. Pollens are abundantly applied on the placenta. In this case also the rate of fertilisation is very slow.
Applications of in vitro Pollination : In plant breeding, the technique of in vitro pollination has lot of potential in at least three different areas,
- overcoming self-incompatibility,
- overcoming cross-incompatibility,
- haploid production through parthenogenesis.
(ii) The technique for protoplast fusion are pritty will defined and highly effective for almost all the systems. Protoplast of desired species/strains are mixed in almost equal proportion; generally they are mixed while still suspended in the enzyme mixture. The protoplast mixture is then subjected to high pH (10.5) and high Ca2+ concentration 50 m molL-1 at 37°C for about 30 min. (high pH Ca2+ treatment). This technique is quite suitable for some species while for some other it may be toxic.
Two types of protoplast fusions are :
Spontaneous protoplast fusion: During isolation of protoplasts for culture, when enzymatic degradation of cell walls is affected, some of the protoplasts, lying in close proximity, may undergo fusion to produce homokarvons or homokaryocytes. each with 2-40 nuclei. The occurrence of multinucleate fusion bodies is more frequent, when protoplasts are prepared from actively dividing cells.
This spontaneous fusion, however, is strictly intraspecific. How ever, spontaneous fusion of protoplasts can also be induced by bringing protoplasts into intimate contact through micromanipulators or micropipettes.
There seems to be a correlation between the size of the leaf and the percentage of protoplasts undergoing spontaneous fusion; protoplasts from young leaves are more likely to undergo this fusion.
Induced protoplast fusion: Somatic hybridization is generally used for fusion of protoplasts either from two different species (inter specific fusion) or from two diverse sources belonging to the same species. To achieve this objective, spontaneous fusion may be of no value, and induced fusion requiring a suitable agent (fusogen) is necessary. In animals, inactivated Sendai virus is needed to induce fusion.
In plants, however, the inducing agent first brings the protoplasts together and then causes them to adhere to one another for bringing about fusion. During the last two decades, a variety of treatments have been successfully utilized for fusion of plant protoplasts. These treatments particularly include the following : NaN02, high pH with high Ca++ ion concentration and polyethylene glycol (PEG).
Applications: Auxin independent growth of hybrids of Nicotiana glauca and A. langsdorffii. Two parental lines cannot produce auxin and thus do not grow on auxin free medium, the hybrid cells produce auxin and are they able to grow and form callus.
(b) (i) Endosperm culture: The endosperm nurses the developing zygotic embryo. For the first time Lamp and Mills (1933) grew7 maize endosperm on nutrient medium 10-20 days after pollination. The endosperm proliferated slightly. In 1949, LaRue succeeded to produce callus from immature endosperm. From India, B.M. Johri and S.S. Bhojwani (1965) at the University of Delhi reported and endosperm culture. Some examples of triploid plants raised from endosperm cultures are : Asparagus officinalis, barley (Hordeum vulgare), rice (Oryza sativa), maize (Zea mays), Prunus persica, Pyrus malus, Citrus gradis, sandle plant (Santalum album).
The triploid plants are self-sterile and usually seedles. This characteristic increases edibility of fruits and desirable in plants such as apple, banana, grape, mulberry, mango, watermelon, etc. These are commercially important edible fruits. The triploids of poplar {Populus tremuloides) have better quality pulpwood. Therefore, it is important to the forest industry,
(ii) Anther culture: When anthers of some plants are cultured on suitable medium to produce haploid plants, it is called anther culture. For the first time S. Guha and P. Maheshwari (1964) produced haploid embryos in vitro from isolated anthers of Datura innoxia. Haploid production has immense use in plant breeding and improvement of crop plants. Haploids provide an easier system for induction of mutation. They can be employed for rapid selection of mutants having traits for disease resistance. The Institute of Crop Breeding and Cultivation (China) has developed the high yielding and blast resistant varieties of rice zhonghua No. 8 and zhonghua No.9 through transfer of desired alien gene.
It is highly useful for the improvement of many crop plants. It is also useful for immediate expression of mutations and quick formation of purelines. This technique was first used in India to produce haploids of Datura. In many plants haploids are also produced by culturing unfertilized ovaries/ovules. In pollen grain culture pollen grains are eseptically removed from the anther and cultured on liquid medium.
(c) The Human Genome Project was an international scientific research project that determined the DNA sequence of the approximately 30,000 — 35,000 genes that make-up the human genome. The greatest achievement of this project is that scientists can find out the structure of human genes and successfully trace the structure of genes. It has been possible to develop the technique regarding the treatment of defective genes only because of the human genome project.
The treatment of the hereditary diseases like cancer has been easier because of this project. DNA interference is a recently developed technique regarding treatment by which it would be possible to treat many incurable diseases. Along with this, the development of human, physical and mental structure etc., are the main achievements of human genome project.
Question 9.
(a) What type of information is obtained from each of the following database. [4]
(i) Taxonomy Browser
(ii) PDB.
(iii) GENSCAN
(iv) PIR.
(b) Explain the method of DNA sequencing using automated DNA sequencing technique. [4]
(c) Write any two examples of each of the following plant hormones commonly used in media preparation: [2]
(i) Auxins.
(ii) Cytokinins.
Answer:
(a) (i) Taxonomy browser: This search tool provides taxonomic information on various species. The Taxonomy database of NCBI has information (including scientific and common names) about all organisms for which some sequence information is available (over 79,000 species).
The server provides genetic information and the taxonomic relationship of the species in question. Taxonomy has links with other servers of NCBI e.g., structure and PubMed.
(ii) PDB (Protein Data Bank) : This database has sequence of those protein whose 3-D structures are known.
Source: NCBI-U.S.A.; EBI, UK.
(iii) Genscan is a notable example of Eukaryotic ab initio gene finders. Genscan is one of the best gene finding algorithms for sequence alignment and gene prediction.
(iv) PIR is a non-redundant annotated protein sequence database, and analytical tools.
(b) Automatic DNA Sequencers: Automatic sequencing machines were developed during 1990’s. It is an improvement of Sanger’s method. In this new method, a different fluorescent dye is tagged to the ddNTP’s. Using this technique, a DNA sequence containing thousands of nucleotides can be determined in a few hours. Each dideoxynucleotide is linked with a fluorescent dye that imparts different colors to all the fragments terminating in that nucleotide. All four labelled ddNTP’s are added to a single capillary tube. It is a refinement of get electrophoresis which separates fastly. DNA fragments of different colors are separated by their size in a single electrophoretic gel.
A current is applied to the gel. The negatively charged DNA strands migrate through the pores of gel towards the positive end. The small sized DNA fragments migrate faster and vice-versa. All fragments of a given length migrate in a single peak. The DNA fragments are illuminated with a laser beam. Then the fluorescent dyes are excited and emit light of specific wavelengths which is recorded by a special ‘recorder’. The DNA sequences are read by determining the sequence of the colours emitted from specific peaks as they pass the detector. This information is fed directly to a computer which determines the sequence. A tracing electrogram of emitted light of the four dyes is generated by the computer. Colour of each dye represents the different nucleotides. Computer converts the data of emitted light in the nucleotide sequences.
(c) (i) IAA (Indole 3-Acetic Acid) and 2, 4-Dichlorophenoxyacetic acid (2, 4-D)
(ii) Kinetin and BAP (Benzyl Amino purine)
